
Marinobacter lutaoensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Marinobacter
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
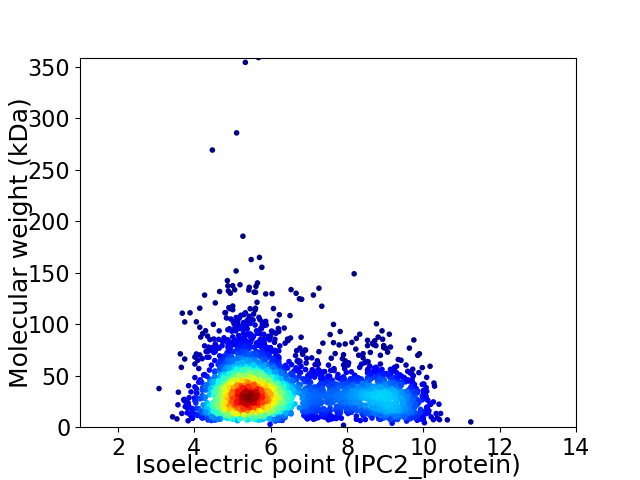
Virtual 2D-PAGE plot for 3411 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V2DR87|A0A1V2DR87_9ALTE Uncharacterized protein OS=Marinobacter lutaoensis OX=135739 GN=BTO32_10035 PE=4 SV=1
MM1 pKa = 7.76KK2 pKa = 10.14NPCVTSRR9 pKa = 11.84WLPLAAVLALAGCGGGSDD27 pKa = 4.16NEE29 pKa = 4.47PGLTSDD35 pKa = 3.49NTGLPEE41 pKa = 4.23GMVEE45 pKa = 4.09KK46 pKa = 9.66TLPAASDD53 pKa = 3.46EE54 pKa = 4.67VYY56 pKa = 11.39VNLEE60 pKa = 3.81TGEE63 pKa = 4.39LVDD66 pKa = 6.4PSDD69 pKa = 3.4TWHH72 pKa = 6.77LKK74 pKa = 10.3ASRR77 pKa = 11.84LTFNLNGGEE86 pKa = 4.21SGTGNVGGALAVAQDD101 pKa = 5.17DD102 pKa = 4.6FYY104 pKa = 11.68KK105 pKa = 11.16DD106 pKa = 3.36NGDD109 pKa = 3.4PDD111 pKa = 4.18EE112 pKa = 4.91NVFTNASASSEE123 pKa = 4.19LEE125 pKa = 3.82HH126 pKa = 8.04LLGPFEE132 pKa = 6.1APAAWQTDD140 pKa = 3.8GLQSAFGSWEE150 pKa = 3.81TWSSYY155 pKa = 11.06DD156 pKa = 3.52YY157 pKa = 10.89STGQISALSDD167 pKa = 2.48IGYY170 pKa = 9.5LVRR173 pKa = 11.84SAEE176 pKa = 4.28GNSFARR182 pKa = 11.84MKK184 pKa = 10.5VVSFDD189 pKa = 3.24FPTRR193 pKa = 11.84ANQGIRR199 pKa = 11.84DD200 pKa = 3.87FEE202 pKa = 4.65FEE204 pKa = 4.47FSVQGAGEE212 pKa = 4.37VAFSDD217 pKa = 3.69QPVTFTPPADD227 pKa = 3.74YY228 pKa = 11.0DD229 pKa = 3.85GGDD232 pKa = 3.12ACFDD236 pKa = 3.78FDD238 pKa = 3.66SGQVVDD244 pKa = 5.52CDD246 pKa = 4.51TSDD249 pKa = 2.96TWDD252 pKa = 3.4VMVGFSGYY260 pKa = 8.37NWYY263 pKa = 10.26LRR265 pKa = 11.84SNSGPSGSGDD275 pKa = 3.2GGASGPIGWDD285 pKa = 3.06EE286 pKa = 4.26LDD288 pKa = 4.36SYY290 pKa = 11.05EE291 pKa = 4.92ADD293 pKa = 3.36PGVPQIYY300 pKa = 10.03AQDD303 pKa = 3.52STGGVFSEE311 pKa = 4.61HH312 pKa = 4.68TWYY315 pKa = 10.52AYY317 pKa = 11.12NLLGNHH323 pKa = 7.02KK324 pKa = 9.35IWPNFRR330 pKa = 11.84TYY332 pKa = 10.8LIKK335 pKa = 10.65ADD337 pKa = 3.58IDD339 pKa = 3.63EE340 pKa = 4.88TEE342 pKa = 3.96ASVWALQIVNYY353 pKa = 10.28YY354 pKa = 10.85DD355 pKa = 4.51DD356 pKa = 6.02DD357 pKa = 4.48GVSGNPTIRR366 pKa = 11.84WHH368 pKa = 7.0RR369 pKa = 11.84VTLNN373 pKa = 2.99
MM1 pKa = 7.76KK2 pKa = 10.14NPCVTSRR9 pKa = 11.84WLPLAAVLALAGCGGGSDD27 pKa = 4.16NEE29 pKa = 4.47PGLTSDD35 pKa = 3.49NTGLPEE41 pKa = 4.23GMVEE45 pKa = 4.09KK46 pKa = 9.66TLPAASDD53 pKa = 3.46EE54 pKa = 4.67VYY56 pKa = 11.39VNLEE60 pKa = 3.81TGEE63 pKa = 4.39LVDD66 pKa = 6.4PSDD69 pKa = 3.4TWHH72 pKa = 6.77LKK74 pKa = 10.3ASRR77 pKa = 11.84LTFNLNGGEE86 pKa = 4.21SGTGNVGGALAVAQDD101 pKa = 5.17DD102 pKa = 4.6FYY104 pKa = 11.68KK105 pKa = 11.16DD106 pKa = 3.36NGDD109 pKa = 3.4PDD111 pKa = 4.18EE112 pKa = 4.91NVFTNASASSEE123 pKa = 4.19LEE125 pKa = 3.82HH126 pKa = 8.04LLGPFEE132 pKa = 6.1APAAWQTDD140 pKa = 3.8GLQSAFGSWEE150 pKa = 3.81TWSSYY155 pKa = 11.06DD156 pKa = 3.52YY157 pKa = 10.89STGQISALSDD167 pKa = 2.48IGYY170 pKa = 9.5LVRR173 pKa = 11.84SAEE176 pKa = 4.28GNSFARR182 pKa = 11.84MKK184 pKa = 10.5VVSFDD189 pKa = 3.24FPTRR193 pKa = 11.84ANQGIRR199 pKa = 11.84DD200 pKa = 3.87FEE202 pKa = 4.65FEE204 pKa = 4.47FSVQGAGEE212 pKa = 4.37VAFSDD217 pKa = 3.69QPVTFTPPADD227 pKa = 3.74YY228 pKa = 11.0DD229 pKa = 3.85GGDD232 pKa = 3.12ACFDD236 pKa = 3.78FDD238 pKa = 3.66SGQVVDD244 pKa = 5.52CDD246 pKa = 4.51TSDD249 pKa = 2.96TWDD252 pKa = 3.4VMVGFSGYY260 pKa = 8.37NWYY263 pKa = 10.26LRR265 pKa = 11.84SNSGPSGSGDD275 pKa = 3.2GGASGPIGWDD285 pKa = 3.06EE286 pKa = 4.26LDD288 pKa = 4.36SYY290 pKa = 11.05EE291 pKa = 4.92ADD293 pKa = 3.36PGVPQIYY300 pKa = 10.03AQDD303 pKa = 3.52STGGVFSEE311 pKa = 4.61HH312 pKa = 4.68TWYY315 pKa = 10.52AYY317 pKa = 11.12NLLGNHH323 pKa = 7.02KK324 pKa = 9.35IWPNFRR330 pKa = 11.84TYY332 pKa = 10.8LIKK335 pKa = 10.65ADD337 pKa = 3.58IDD339 pKa = 3.63EE340 pKa = 4.88TEE342 pKa = 3.96ASVWALQIVNYY353 pKa = 10.28YY354 pKa = 10.85DD355 pKa = 4.51DD356 pKa = 6.02DD357 pKa = 4.48GVSGNPTIRR366 pKa = 11.84WHH368 pKa = 7.0RR369 pKa = 11.84VTLNN373 pKa = 2.99
Molecular weight: 40.23 kDa
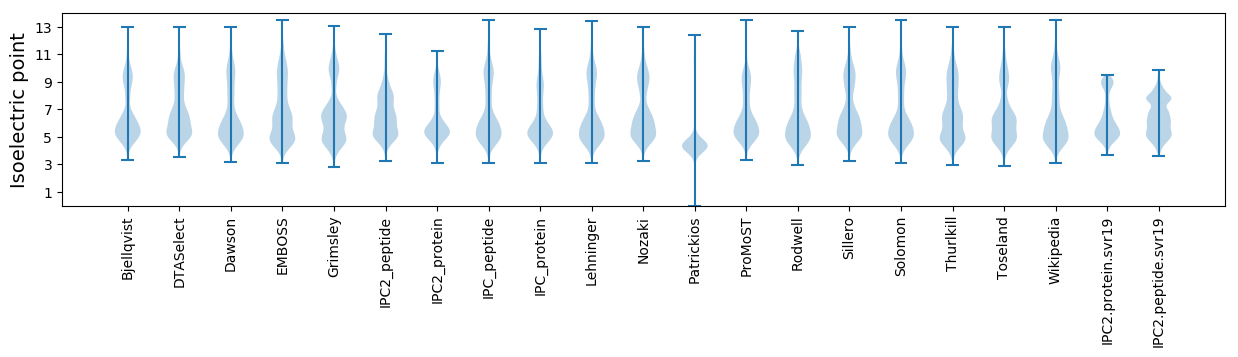
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V2DUZ4|A0A1V2DUZ4_9ALTE Poly(Hydroxyalkanoate) granule-associated protein OS=Marinobacter lutaoensis OX=135739 GN=BTO32_07740 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.91VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.44GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.91VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.44GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1153080 |
15 |
3247 |
338.0 |
37.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.666 ± 0.045 | 0.932 ± 0.012 |
5.807 ± 0.035 | 6.297 ± 0.042 |
3.553 ± 0.023 | 7.915 ± 0.041 |
2.3 ± 0.021 | 4.728 ± 0.035 |
2.983 ± 0.035 | 11.24 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.319 ± 0.019 | 2.776 ± 0.023 |
5.118 ± 0.028 | 3.995 ± 0.027 |
7.613 ± 0.041 | 5.259 ± 0.032 |
5.134 ± 0.03 | 7.4 ± 0.034 |
1.413 ± 0.018 | 2.552 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
