
Purpureocillium lilacinum (Paecilomyces lilacinus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Ophiocordycipitaceae; Purpureocillium
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
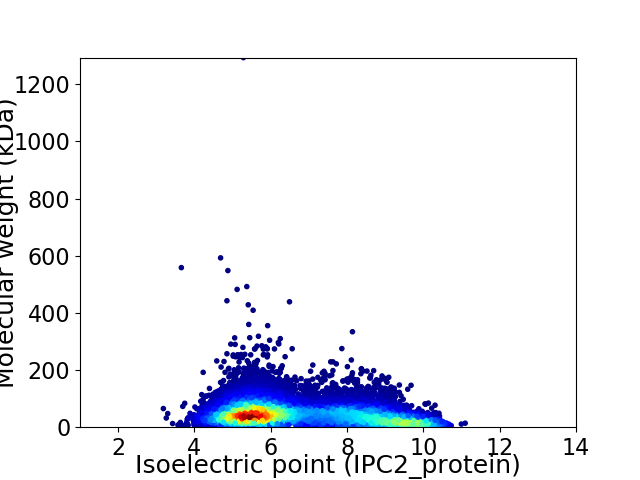
Virtual 2D-PAGE plot for 11750 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
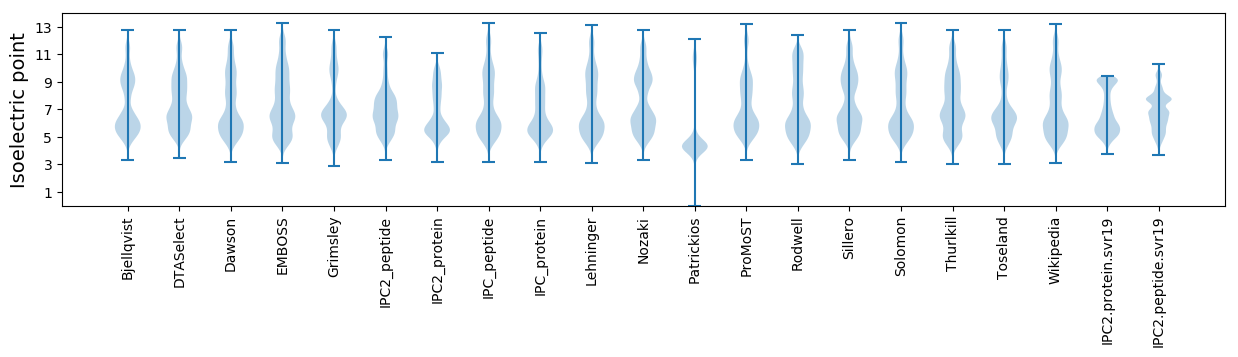
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A179H7P0|A0A179H7P0_PURLI GTPase activating protein OS=Purpureocillium lilacinum OX=33203 GN=VFPBJ_00237 PE=4 SV=1
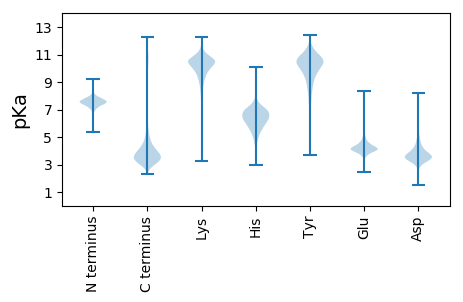
MM1 pKa = 7.52PRR3 pKa = 11.84VTSDD7 pKa = 3.13SFSGEE12 pKa = 3.99SAEE15 pKa = 4.22VQRR18 pKa = 11.84DD19 pKa = 3.24QALATKK25 pKa = 10.53LDD27 pKa = 3.74DD28 pKa = 4.69EE29 pKa = 4.72YY30 pKa = 11.65SEE32 pKa = 5.2KK33 pKa = 11.03YY34 pKa = 10.58LDD36 pKa = 4.17GLTILLDD43 pKa = 3.6YY44 pKa = 11.16LNAGNTYY51 pKa = 11.19DD52 pKa = 4.75DD53 pKa = 4.35MLRR56 pKa = 11.84VIPDD60 pKa = 3.17AQEE63 pKa = 3.25WCRR66 pKa = 11.84EE67 pKa = 4.1YY68 pKa = 11.06FSGRR72 pKa = 11.84MVAPNRR78 pKa = 11.84LNEE81 pKa = 4.36EE82 pKa = 4.0VLFWHH87 pKa = 7.08PNLLDD92 pKa = 3.79LSLPSWRR99 pKa = 11.84RR100 pKa = 11.84LDD102 pKa = 3.75GPNNFDD108 pKa = 3.25EE109 pKa = 4.63TQLFGDD115 pKa = 4.41VDD117 pKa = 3.63PMEE120 pKa = 4.51GFEE123 pKa = 5.88YY124 pKa = 10.37PLTWEE129 pKa = 4.7SEE131 pKa = 4.2DD132 pKa = 3.37EE133 pKa = 4.34TLVVEE138 pKa = 5.14MDD140 pKa = 2.96QHH142 pKa = 6.23MNDD145 pKa = 3.42DD146 pKa = 3.74VYY148 pKa = 11.57AFEE151 pKa = 5.75LGQHH155 pKa = 6.0VDD157 pKa = 2.71NDD159 pKa = 3.97YY160 pKa = 11.61NFAEE164 pKa = 4.45AEE166 pKa = 4.11MGHH169 pKa = 5.26VAEE172 pKa = 5.56AVFSDD177 pKa = 4.01GGYY180 pKa = 10.15QINGDD185 pKa = 4.25GISDD189 pKa = 4.06NLDD192 pKa = 3.38LDD194 pKa = 4.13AVLPIYY200 pKa = 10.27GAISEE205 pKa = 4.59WVPFSDD211 pKa = 5.31DD212 pKa = 3.42EE213 pKa = 4.38VHH215 pKa = 7.21AFLDD219 pKa = 3.79RR220 pKa = 11.84HH221 pKa = 4.86SHH223 pKa = 4.67RR224 pKa = 11.84TYY226 pKa = 11.32EE227 pKa = 3.88NLLAGGHH234 pKa = 6.62DD235 pKa = 3.78VLFDD239 pKa = 4.62DD240 pKa = 4.92YY241 pKa = 11.57LFLRR245 pKa = 11.84GAPLMASDD253 pKa = 4.26LTRR256 pKa = 11.84AVAYY260 pKa = 8.35NLPLILSCLASIPLHH275 pKa = 5.66EE276 pKa = 4.62VVDD279 pKa = 4.58LDD281 pKa = 3.93VLGMEE286 pKa = 4.49TDD288 pKa = 4.19DD289 pKa = 5.36NEE291 pKa = 4.58STT293 pKa = 3.81
MM1 pKa = 7.52PRR3 pKa = 11.84VTSDD7 pKa = 3.13SFSGEE12 pKa = 3.99SAEE15 pKa = 4.22VQRR18 pKa = 11.84DD19 pKa = 3.24QALATKK25 pKa = 10.53LDD27 pKa = 3.74DD28 pKa = 4.69EE29 pKa = 4.72YY30 pKa = 11.65SEE32 pKa = 5.2KK33 pKa = 11.03YY34 pKa = 10.58LDD36 pKa = 4.17GLTILLDD43 pKa = 3.6YY44 pKa = 11.16LNAGNTYY51 pKa = 11.19DD52 pKa = 4.75DD53 pKa = 4.35MLRR56 pKa = 11.84VIPDD60 pKa = 3.17AQEE63 pKa = 3.25WCRR66 pKa = 11.84EE67 pKa = 4.1YY68 pKa = 11.06FSGRR72 pKa = 11.84MVAPNRR78 pKa = 11.84LNEE81 pKa = 4.36EE82 pKa = 4.0VLFWHH87 pKa = 7.08PNLLDD92 pKa = 3.79LSLPSWRR99 pKa = 11.84RR100 pKa = 11.84LDD102 pKa = 3.75GPNNFDD108 pKa = 3.25EE109 pKa = 4.63TQLFGDD115 pKa = 4.41VDD117 pKa = 3.63PMEE120 pKa = 4.51GFEE123 pKa = 5.88YY124 pKa = 10.37PLTWEE129 pKa = 4.7SEE131 pKa = 4.2DD132 pKa = 3.37EE133 pKa = 4.34TLVVEE138 pKa = 5.14MDD140 pKa = 2.96QHH142 pKa = 6.23MNDD145 pKa = 3.42DD146 pKa = 3.74VYY148 pKa = 11.57AFEE151 pKa = 5.75LGQHH155 pKa = 6.0VDD157 pKa = 2.71NDD159 pKa = 3.97YY160 pKa = 11.61NFAEE164 pKa = 4.45AEE166 pKa = 4.11MGHH169 pKa = 5.26VAEE172 pKa = 5.56AVFSDD177 pKa = 4.01GGYY180 pKa = 10.15QINGDD185 pKa = 4.25GISDD189 pKa = 4.06NLDD192 pKa = 3.38LDD194 pKa = 4.13AVLPIYY200 pKa = 10.27GAISEE205 pKa = 4.59WVPFSDD211 pKa = 5.31DD212 pKa = 3.42EE213 pKa = 4.38VHH215 pKa = 7.21AFLDD219 pKa = 3.79RR220 pKa = 11.84HH221 pKa = 4.86SHH223 pKa = 4.67RR224 pKa = 11.84TYY226 pKa = 11.32EE227 pKa = 3.88NLLAGGHH234 pKa = 6.62DD235 pKa = 3.78VLFDD239 pKa = 4.62DD240 pKa = 4.92YY241 pKa = 11.57LFLRR245 pKa = 11.84GAPLMASDD253 pKa = 4.26LTRR256 pKa = 11.84AVAYY260 pKa = 8.35NLPLILSCLASIPLHH275 pKa = 5.66EE276 pKa = 4.62VVDD279 pKa = 4.58LDD281 pKa = 3.93VLGMEE286 pKa = 4.49TDD288 pKa = 4.19DD289 pKa = 5.36NEE291 pKa = 4.58STT293 pKa = 3.81
Molecular weight: 33.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|A0A179GC70|LCSQ_PURLI Nucleotidyltransferase lcsQ OS=Purpureocillium lilacinum OX=33203 GN=lcsQ PE=2 SV=1
MM1 pKa = 7.38FSGRR5 pKa = 11.84RR6 pKa = 11.84TRR8 pKa = 11.84RR9 pKa = 11.84ANAHH13 pKa = 5.74HH14 pKa = 6.57HH15 pKa = 5.87HH16 pKa = 6.6HH17 pKa = 6.58HH18 pKa = 6.41HH19 pKa = 5.98TTTTTTRR26 pKa = 11.84APRR29 pKa = 11.84RR30 pKa = 11.84SIFSRR35 pKa = 11.84RR36 pKa = 11.84QRR38 pKa = 11.84VAVHH42 pKa = 4.84HH43 pKa = 5.95QKK45 pKa = 10.56RR46 pKa = 11.84KK47 pKa = 8.39PTMKK51 pKa = 10.55DD52 pKa = 3.11KK53 pKa = 11.05ISGALTKK60 pKa = 10.7LRR62 pKa = 11.84GSLTRR67 pKa = 11.84RR68 pKa = 11.84PGLKK72 pKa = 10.1AAGTRR77 pKa = 11.84RR78 pKa = 11.84MHH80 pKa = 5.68GTDD83 pKa = 2.72GRR85 pKa = 11.84GSHH88 pKa = 6.15RR89 pKa = 11.84AGRR92 pKa = 11.84RR93 pKa = 11.84FFF95 pKa = 4.56
MM1 pKa = 7.38FSGRR5 pKa = 11.84RR6 pKa = 11.84TRR8 pKa = 11.84RR9 pKa = 11.84ANAHH13 pKa = 5.74HH14 pKa = 6.57HH15 pKa = 5.87HH16 pKa = 6.6HH17 pKa = 6.58HH18 pKa = 6.41HH19 pKa = 5.98TTTTTTRR26 pKa = 11.84APRR29 pKa = 11.84RR30 pKa = 11.84SIFSRR35 pKa = 11.84RR36 pKa = 11.84QRR38 pKa = 11.84VAVHH42 pKa = 4.84HH43 pKa = 5.95QKK45 pKa = 10.56RR46 pKa = 11.84KK47 pKa = 8.39PTMKK51 pKa = 10.55DD52 pKa = 3.11KK53 pKa = 11.05ISGALTKK60 pKa = 10.7LRR62 pKa = 11.84GSLTRR67 pKa = 11.84RR68 pKa = 11.84PGLKK72 pKa = 10.1AAGTRR77 pKa = 11.84RR78 pKa = 11.84MHH80 pKa = 5.68GTDD83 pKa = 2.72GRR85 pKa = 11.84GSHH88 pKa = 6.15RR89 pKa = 11.84AGRR92 pKa = 11.84RR93 pKa = 11.84FFF95 pKa = 4.56
Molecular weight: 10.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5417339 |
45 |
11859 |
461.1 |
50.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.846 ± 0.023 | 1.34 ± 0.01 |
5.902 ± 0.017 | 5.789 ± 0.023 |
3.529 ± 0.013 | 7.433 ± 0.02 |
2.441 ± 0.012 | 4.224 ± 0.016 |
4.445 ± 0.02 | 8.74 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.221 ± 0.008 | 3.218 ± 0.014 |
6.2 ± 0.027 | 3.928 ± 0.019 |
6.687 ± 0.023 | 7.832 ± 0.025 |
5.744 ± 0.017 | 6.394 ± 0.016 |
1.525 ± 0.009 | 2.552 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
