
Hydrogenovibrio marinus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Thiotrichales; Piscirickettsiaceae; Hydrogenovibrio
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
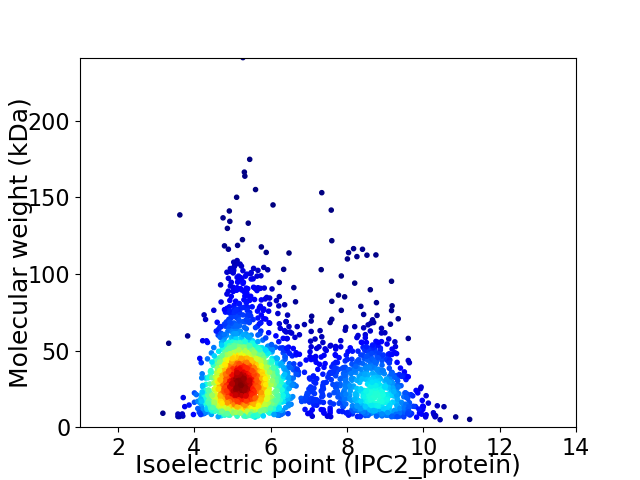
Virtual 2D-PAGE plot for 2330 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A067A105|A0A067A105_HYDMR Pribosyltran domain-containing protein OS=Hydrogenovibrio marinus OX=28885 GN=EI16_08415 PE=4 SV=1
MM1 pKa = 7.04ITNITDD7 pKa = 3.49TNGDD11 pKa = 3.6YY12 pKa = 11.43SNVILHH18 pKa = 5.88GTGEE22 pKa = 4.17PGATVTLLSRR32 pKa = 11.84EE33 pKa = 3.99GSTTGGNDD41 pKa = 3.12TGSWTYY47 pKa = 9.79TAVEE51 pKa = 4.19TSTPIVVDD59 pKa = 4.36ANGNWTVDD67 pKa = 2.99ISNLPNTPIDD77 pKa = 3.84DD78 pKa = 4.09NEE80 pKa = 4.23FFKK83 pKa = 10.62ATQIDD88 pKa = 3.5ASGNVSADD96 pKa = 3.42SDD98 pKa = 4.35SVHH101 pKa = 5.94YY102 pKa = 9.07WHH104 pKa = 7.55GDD106 pKa = 2.85WSSVATEE113 pKa = 4.26ANDD116 pKa = 3.82DD117 pKa = 3.92YY118 pKa = 12.08VMMGSGKK125 pKa = 9.79DD126 pKa = 3.33QIIISSNDD134 pKa = 3.44TNDD137 pKa = 3.11HH138 pKa = 5.69FVVDD142 pKa = 4.32GGDD145 pKa = 3.89NIDD148 pKa = 3.51TAVFSGKK155 pKa = 10.18FSDD158 pKa = 4.35YY159 pKa = 11.43ALSTSSTGSLIVTEE173 pKa = 4.9GVSTDD178 pKa = 3.3SNGNGVGDD186 pKa = 3.62VDD188 pKa = 3.69EE189 pKa = 5.02LRR191 pKa = 11.84NIEE194 pKa = 4.27KK195 pKa = 10.83VKK197 pKa = 9.85FTDD200 pKa = 3.36GTYY203 pKa = 10.64DD204 pKa = 3.18VPTGVFKK211 pKa = 11.43LNGISVDD218 pKa = 3.52IEE220 pKa = 4.43HH221 pKa = 7.33DD222 pKa = 3.55TGTSSTDD229 pKa = 3.23RR230 pKa = 11.84ITNDD234 pKa = 2.56STLKK238 pKa = 9.37VTGLVSGATIEE249 pKa = 4.14YY250 pKa = 10.35SSDD253 pKa = 3.25GGQTWTTQYY262 pKa = 11.28NPQEE266 pKa = 4.2GVNNISVRR274 pKa = 11.84QVDD277 pKa = 4.24AEE279 pKa = 4.25GHH281 pKa = 5.86TSDD284 pKa = 4.25SSNVSFTLDD293 pKa = 3.24TQAGANIDD301 pKa = 3.65VDD303 pKa = 3.6ILTGDD308 pKa = 4.2GYY310 pKa = 10.06LTAAEE315 pKa = 4.58AFPGVKK321 pKa = 10.07VPVTGYY327 pKa = 10.11VQGDD331 pKa = 4.0AQPGDD336 pKa = 4.12TISIILDD343 pKa = 3.38GNVIGTGTVSSDD355 pKa = 3.46VNSSGAYY362 pKa = 7.39TFSVDD367 pKa = 4.35VLGSDD372 pKa = 3.78LADD375 pKa = 3.78GSGMIPEE382 pKa = 5.04LKK384 pKa = 10.02ATVTGSDD391 pKa = 3.29VAGNTFSAEE400 pKa = 3.98STEE403 pKa = 4.79VYY405 pKa = 9.95MKK407 pKa = 10.94DD408 pKa = 4.12FIATVSINVTDD419 pKa = 4.52GNTSDD424 pKa = 3.6SDD426 pKa = 3.95NVISASEE433 pKa = 3.87ASSTVVSGNVEE444 pKa = 3.6VGGQVSSITISDD456 pKa = 3.69GTNTLTIPANQITVYY471 pKa = 10.9SNGYY475 pKa = 7.37YY476 pKa = 9.38TVNTDD481 pKa = 3.29VSSLNDD487 pKa = 3.5GVLTVSVEE495 pKa = 4.21ASDD498 pKa = 3.83KK499 pKa = 11.02LGNVGTNSTTIEE511 pKa = 4.71KK512 pKa = 8.78DD513 pKa = 3.18TQAQAGTVTVDD524 pKa = 4.87SITDD528 pKa = 3.64DD529 pKa = 4.31NIVNTMEE536 pKa = 4.25SGQTIAVTGTATGGDD551 pKa = 3.54ISSGDD556 pKa = 3.94TLTMTINGKK565 pKa = 10.19DD566 pKa = 3.36YY567 pKa = 10.35STTVNSDD574 pKa = 3.51GNWSVDD580 pKa = 3.45VAGSDD585 pKa = 3.97LAADD589 pKa = 3.81ADD591 pKa = 3.97KK592 pKa = 11.56GFTASVASSDD602 pKa = 3.53AAGNTVTSSAAHH614 pKa = 6.71SYY616 pKa = 9.41TVDD619 pKa = 3.58SQPEE623 pKa = 4.24GHH625 pKa = 7.44DD626 pKa = 3.34EE627 pKa = 4.11TQTMDD632 pKa = 3.64FEE634 pKa = 4.84SATVSQKK641 pKa = 6.25TTNVVITLDD650 pKa = 3.3ISGSMDD656 pKa = 3.29SDD658 pKa = 3.7SRR660 pKa = 11.84LALAKK665 pKa = 10.37SALEE669 pKa = 3.96KK670 pKa = 10.34MINAYY675 pKa = 10.28DD676 pKa = 3.54NQGNVNVKK684 pKa = 9.92LVAFNGGATLEE695 pKa = 4.11QHH697 pKa = 7.15DD698 pKa = 4.14GSVWLKK704 pKa = 10.29ADD706 pKa = 3.62EE707 pKa = 5.27AISIIKK713 pKa = 9.71DD714 pKa = 3.49LQAGGSTNYY723 pKa = 10.09EE724 pKa = 3.9DD725 pKa = 5.71AVLKK729 pKa = 10.06TYY731 pKa = 11.2SNYY734 pKa = 10.11SEE736 pKa = 4.02PTADD740 pKa = 2.96KK741 pKa = 9.75TVAYY745 pKa = 8.72FISDD749 pKa = 4.15GEE751 pKa = 4.39PTTEE755 pKa = 3.8NYY757 pKa = 10.22GGNGSGTAGGFLDD770 pKa = 4.36NDD772 pKa = 4.32YY773 pKa = 11.58EE774 pKa = 4.48HH775 pKa = 6.99GWDD778 pKa = 3.62NFVSQYY784 pKa = 11.01VDD786 pKa = 3.54DD787 pKa = 4.41LHH789 pKa = 8.49VVALGDD795 pKa = 3.58DD796 pKa = 4.0ALNSKK801 pKa = 10.15YY802 pKa = 10.86LDD804 pKa = 3.59VLASAGNNVSTVTEE818 pKa = 4.09VSDD821 pKa = 4.5ASQLSDD827 pKa = 5.12AIVPTNTEE835 pKa = 3.84LSVTGTVSDD844 pKa = 4.09NVTGGDD850 pKa = 3.72GAITIQSITVDD861 pKa = 3.34GTAYY865 pKa = 9.67TSSTFPSEE873 pKa = 3.72GVAINGKK880 pKa = 8.33GTLAFDD886 pKa = 4.76FSTGNYY892 pKa = 7.53TYY894 pKa = 10.93SAGSNEE900 pKa = 3.81FTKK903 pKa = 10.74DD904 pKa = 3.19VTQMFTVTAADD915 pKa = 3.83ADD917 pKa = 4.15GDD919 pKa = 4.1TADD922 pKa = 3.74VKK924 pKa = 11.29VSINVNVDD932 pKa = 3.24DD933 pKa = 4.25TASNATLSMSIGDD946 pKa = 4.06AEE948 pKa = 4.49VTTTHH953 pKa = 6.74NYY955 pKa = 9.96TYY957 pKa = 11.02DD958 pKa = 3.42SDD960 pKa = 3.97DD961 pKa = 3.63ARR963 pKa = 11.84NANHH967 pKa = 7.26DD968 pKa = 4.21NYY970 pKa = 10.99HH971 pKa = 6.11SLHH974 pKa = 7.03DD975 pKa = 4.43ALEE978 pKa = 4.48GQNGCNHH985 pKa = 6.29SNGTLVYY992 pKa = 10.42KK993 pKa = 10.57NVNEE997 pKa = 4.33TSWNLSDD1004 pKa = 5.79SSDD1007 pKa = 3.36KK1008 pKa = 11.24LFAITGNSHH1017 pKa = 6.06NANINLSGGNNTLIFEE1033 pKa = 5.01KK1034 pKa = 10.89DD1035 pKa = 3.23PGSNIAVNFGSGKK1048 pKa = 10.25DD1049 pKa = 3.63VLILPGSEE1057 pKa = 3.58SDD1059 pKa = 3.86YY1060 pKa = 11.18NLSAFHH1066 pKa = 6.79NNGGVLSGQITGHH1079 pKa = 6.94DD1080 pKa = 3.63NMNLTVNNLDD1090 pKa = 3.84CLVFSDD1096 pKa = 4.64QVMGDD1101 pKa = 3.06SSLYY1105 pKa = 10.38KK1106 pKa = 10.82GEE1108 pKa = 4.17TSNHH1112 pKa = 3.83YY1113 pKa = 9.61TYY1115 pKa = 11.16DD1116 pKa = 3.73LSASASLTDD1125 pKa = 3.53TDD1127 pKa = 4.29GSEE1130 pKa = 4.18SLSDD1134 pKa = 3.28ITISGLPDD1142 pKa = 4.5DD1143 pKa = 5.35GSVTVTGTDD1152 pKa = 3.04VSKK1155 pKa = 11.49NDD1157 pKa = 3.51DD1158 pKa = 2.97GTYY1161 pKa = 10.3NVKK1164 pKa = 10.46LQDD1167 pKa = 4.01DD1168 pKa = 4.44GKK1170 pKa = 10.71IADD1173 pKa = 4.23DD1174 pKa = 3.97VKK1176 pKa = 10.72ISSSKK1181 pKa = 10.54EE1182 pKa = 3.82LTSSEE1187 pKa = 4.36LNDD1190 pKa = 3.01VHH1192 pKa = 8.98ASVTSTEE1199 pKa = 4.41SNGNEE1204 pKa = 3.96TATTEE1209 pKa = 4.21VNEE1212 pKa = 4.4SGDD1215 pKa = 3.49NFLYY1219 pKa = 10.86ADD1221 pKa = 3.84TGDD1224 pKa = 4.08DD1225 pKa = 4.12LFVGTNQSDD1234 pKa = 4.12TIKK1237 pKa = 10.66VDD1239 pKa = 3.33GDD1241 pKa = 3.62GSTTVQNFDD1250 pKa = 3.37VQNDD1254 pKa = 3.64VLDD1257 pKa = 4.52LSDD1260 pKa = 5.48VIADD1264 pKa = 4.01DD1265 pKa = 4.21QVVTQDD1271 pKa = 3.62TLSQYY1276 pKa = 9.97LTVTQTDD1283 pKa = 3.14KK1284 pKa = 10.71GTEE1287 pKa = 3.96VTVDD1291 pKa = 3.47SNGKK1295 pKa = 9.26DD1296 pKa = 3.38VAGGEE1301 pKa = 4.37VANIMLEE1308 pKa = 3.91GLNDD1312 pKa = 3.98SNNLQIQIDD1321 pKa = 4.07DD1322 pKa = 3.99NKK1324 pKa = 10.44VDD1326 pKa = 3.52YY1327 pKa = 11.12HH1328 pKa = 6.78DD1329 pKa = 3.94
MM1 pKa = 7.04ITNITDD7 pKa = 3.49TNGDD11 pKa = 3.6YY12 pKa = 11.43SNVILHH18 pKa = 5.88GTGEE22 pKa = 4.17PGATVTLLSRR32 pKa = 11.84EE33 pKa = 3.99GSTTGGNDD41 pKa = 3.12TGSWTYY47 pKa = 9.79TAVEE51 pKa = 4.19TSTPIVVDD59 pKa = 4.36ANGNWTVDD67 pKa = 2.99ISNLPNTPIDD77 pKa = 3.84DD78 pKa = 4.09NEE80 pKa = 4.23FFKK83 pKa = 10.62ATQIDD88 pKa = 3.5ASGNVSADD96 pKa = 3.42SDD98 pKa = 4.35SVHH101 pKa = 5.94YY102 pKa = 9.07WHH104 pKa = 7.55GDD106 pKa = 2.85WSSVATEE113 pKa = 4.26ANDD116 pKa = 3.82DD117 pKa = 3.92YY118 pKa = 12.08VMMGSGKK125 pKa = 9.79DD126 pKa = 3.33QIIISSNDD134 pKa = 3.44TNDD137 pKa = 3.11HH138 pKa = 5.69FVVDD142 pKa = 4.32GGDD145 pKa = 3.89NIDD148 pKa = 3.51TAVFSGKK155 pKa = 10.18FSDD158 pKa = 4.35YY159 pKa = 11.43ALSTSSTGSLIVTEE173 pKa = 4.9GVSTDD178 pKa = 3.3SNGNGVGDD186 pKa = 3.62VDD188 pKa = 3.69EE189 pKa = 5.02LRR191 pKa = 11.84NIEE194 pKa = 4.27KK195 pKa = 10.83VKK197 pKa = 9.85FTDD200 pKa = 3.36GTYY203 pKa = 10.64DD204 pKa = 3.18VPTGVFKK211 pKa = 11.43LNGISVDD218 pKa = 3.52IEE220 pKa = 4.43HH221 pKa = 7.33DD222 pKa = 3.55TGTSSTDD229 pKa = 3.23RR230 pKa = 11.84ITNDD234 pKa = 2.56STLKK238 pKa = 9.37VTGLVSGATIEE249 pKa = 4.14YY250 pKa = 10.35SSDD253 pKa = 3.25GGQTWTTQYY262 pKa = 11.28NPQEE266 pKa = 4.2GVNNISVRR274 pKa = 11.84QVDD277 pKa = 4.24AEE279 pKa = 4.25GHH281 pKa = 5.86TSDD284 pKa = 4.25SSNVSFTLDD293 pKa = 3.24TQAGANIDD301 pKa = 3.65VDD303 pKa = 3.6ILTGDD308 pKa = 4.2GYY310 pKa = 10.06LTAAEE315 pKa = 4.58AFPGVKK321 pKa = 10.07VPVTGYY327 pKa = 10.11VQGDD331 pKa = 4.0AQPGDD336 pKa = 4.12TISIILDD343 pKa = 3.38GNVIGTGTVSSDD355 pKa = 3.46VNSSGAYY362 pKa = 7.39TFSVDD367 pKa = 4.35VLGSDD372 pKa = 3.78LADD375 pKa = 3.78GSGMIPEE382 pKa = 5.04LKK384 pKa = 10.02ATVTGSDD391 pKa = 3.29VAGNTFSAEE400 pKa = 3.98STEE403 pKa = 4.79VYY405 pKa = 9.95MKK407 pKa = 10.94DD408 pKa = 4.12FIATVSINVTDD419 pKa = 4.52GNTSDD424 pKa = 3.6SDD426 pKa = 3.95NVISASEE433 pKa = 3.87ASSTVVSGNVEE444 pKa = 3.6VGGQVSSITISDD456 pKa = 3.69GTNTLTIPANQITVYY471 pKa = 10.9SNGYY475 pKa = 7.37YY476 pKa = 9.38TVNTDD481 pKa = 3.29VSSLNDD487 pKa = 3.5GVLTVSVEE495 pKa = 4.21ASDD498 pKa = 3.83KK499 pKa = 11.02LGNVGTNSTTIEE511 pKa = 4.71KK512 pKa = 8.78DD513 pKa = 3.18TQAQAGTVTVDD524 pKa = 4.87SITDD528 pKa = 3.64DD529 pKa = 4.31NIVNTMEE536 pKa = 4.25SGQTIAVTGTATGGDD551 pKa = 3.54ISSGDD556 pKa = 3.94TLTMTINGKK565 pKa = 10.19DD566 pKa = 3.36YY567 pKa = 10.35STTVNSDD574 pKa = 3.51GNWSVDD580 pKa = 3.45VAGSDD585 pKa = 3.97LAADD589 pKa = 3.81ADD591 pKa = 3.97KK592 pKa = 11.56GFTASVASSDD602 pKa = 3.53AAGNTVTSSAAHH614 pKa = 6.71SYY616 pKa = 9.41TVDD619 pKa = 3.58SQPEE623 pKa = 4.24GHH625 pKa = 7.44DD626 pKa = 3.34EE627 pKa = 4.11TQTMDD632 pKa = 3.64FEE634 pKa = 4.84SATVSQKK641 pKa = 6.25TTNVVITLDD650 pKa = 3.3ISGSMDD656 pKa = 3.29SDD658 pKa = 3.7SRR660 pKa = 11.84LALAKK665 pKa = 10.37SALEE669 pKa = 3.96KK670 pKa = 10.34MINAYY675 pKa = 10.28DD676 pKa = 3.54NQGNVNVKK684 pKa = 9.92LVAFNGGATLEE695 pKa = 4.11QHH697 pKa = 7.15DD698 pKa = 4.14GSVWLKK704 pKa = 10.29ADD706 pKa = 3.62EE707 pKa = 5.27AISIIKK713 pKa = 9.71DD714 pKa = 3.49LQAGGSTNYY723 pKa = 10.09EE724 pKa = 3.9DD725 pKa = 5.71AVLKK729 pKa = 10.06TYY731 pKa = 11.2SNYY734 pKa = 10.11SEE736 pKa = 4.02PTADD740 pKa = 2.96KK741 pKa = 9.75TVAYY745 pKa = 8.72FISDD749 pKa = 4.15GEE751 pKa = 4.39PTTEE755 pKa = 3.8NYY757 pKa = 10.22GGNGSGTAGGFLDD770 pKa = 4.36NDD772 pKa = 4.32YY773 pKa = 11.58EE774 pKa = 4.48HH775 pKa = 6.99GWDD778 pKa = 3.62NFVSQYY784 pKa = 11.01VDD786 pKa = 3.54DD787 pKa = 4.41LHH789 pKa = 8.49VVALGDD795 pKa = 3.58DD796 pKa = 4.0ALNSKK801 pKa = 10.15YY802 pKa = 10.86LDD804 pKa = 3.59VLASAGNNVSTVTEE818 pKa = 4.09VSDD821 pKa = 4.5ASQLSDD827 pKa = 5.12AIVPTNTEE835 pKa = 3.84LSVTGTVSDD844 pKa = 4.09NVTGGDD850 pKa = 3.72GAITIQSITVDD861 pKa = 3.34GTAYY865 pKa = 9.67TSSTFPSEE873 pKa = 3.72GVAINGKK880 pKa = 8.33GTLAFDD886 pKa = 4.76FSTGNYY892 pKa = 7.53TYY894 pKa = 10.93SAGSNEE900 pKa = 3.81FTKK903 pKa = 10.74DD904 pKa = 3.19VTQMFTVTAADD915 pKa = 3.83ADD917 pKa = 4.15GDD919 pKa = 4.1TADD922 pKa = 3.74VKK924 pKa = 11.29VSINVNVDD932 pKa = 3.24DD933 pKa = 4.25TASNATLSMSIGDD946 pKa = 4.06AEE948 pKa = 4.49VTTTHH953 pKa = 6.74NYY955 pKa = 9.96TYY957 pKa = 11.02DD958 pKa = 3.42SDD960 pKa = 3.97DD961 pKa = 3.63ARR963 pKa = 11.84NANHH967 pKa = 7.26DD968 pKa = 4.21NYY970 pKa = 10.99HH971 pKa = 6.11SLHH974 pKa = 7.03DD975 pKa = 4.43ALEE978 pKa = 4.48GQNGCNHH985 pKa = 6.29SNGTLVYY992 pKa = 10.42KK993 pKa = 10.57NVNEE997 pKa = 4.33TSWNLSDD1004 pKa = 5.79SSDD1007 pKa = 3.36KK1008 pKa = 11.24LFAITGNSHH1017 pKa = 6.06NANINLSGGNNTLIFEE1033 pKa = 5.01KK1034 pKa = 10.89DD1035 pKa = 3.23PGSNIAVNFGSGKK1048 pKa = 10.25DD1049 pKa = 3.63VLILPGSEE1057 pKa = 3.58SDD1059 pKa = 3.86YY1060 pKa = 11.18NLSAFHH1066 pKa = 6.79NNGGVLSGQITGHH1079 pKa = 6.94DD1080 pKa = 3.63NMNLTVNNLDD1090 pKa = 3.84CLVFSDD1096 pKa = 4.64QVMGDD1101 pKa = 3.06SSLYY1105 pKa = 10.38KK1106 pKa = 10.82GEE1108 pKa = 4.17TSNHH1112 pKa = 3.83YY1113 pKa = 9.61TYY1115 pKa = 11.16DD1116 pKa = 3.73LSASASLTDD1125 pKa = 3.53TDD1127 pKa = 4.29GSEE1130 pKa = 4.18SLSDD1134 pKa = 3.28ITISGLPDD1142 pKa = 4.5DD1143 pKa = 5.35GSVTVTGTDD1152 pKa = 3.04VSKK1155 pKa = 11.49NDD1157 pKa = 3.51DD1158 pKa = 2.97GTYY1161 pKa = 10.3NVKK1164 pKa = 10.46LQDD1167 pKa = 4.01DD1168 pKa = 4.44GKK1170 pKa = 10.71IADD1173 pKa = 4.23DD1174 pKa = 3.97VKK1176 pKa = 10.72ISSSKK1181 pKa = 10.54EE1182 pKa = 3.82LTSSEE1187 pKa = 4.36LNDD1190 pKa = 3.01VHH1192 pKa = 8.98ASVTSTEE1199 pKa = 4.41SNGNEE1204 pKa = 3.96TATTEE1209 pKa = 4.21VNEE1212 pKa = 4.4SGDD1215 pKa = 3.49NFLYY1219 pKa = 10.86ADD1221 pKa = 3.84TGDD1224 pKa = 4.08DD1225 pKa = 4.12LFVGTNQSDD1234 pKa = 4.12TIKK1237 pKa = 10.66VDD1239 pKa = 3.33GDD1241 pKa = 3.62GSTTVQNFDD1250 pKa = 3.37VQNDD1254 pKa = 3.64VLDD1257 pKa = 4.52LSDD1260 pKa = 5.48VIADD1264 pKa = 4.01DD1265 pKa = 4.21QVVTQDD1271 pKa = 3.62TLSQYY1276 pKa = 9.97LTVTQTDD1283 pKa = 3.14KK1284 pKa = 10.71GTEE1287 pKa = 3.96VTVDD1291 pKa = 3.47SNGKK1295 pKa = 9.26DD1296 pKa = 3.38VAGGEE1301 pKa = 4.37VANIMLEE1308 pKa = 3.91GLNDD1312 pKa = 3.98SNNLQIQIDD1321 pKa = 4.07DD1322 pKa = 3.99NKK1324 pKa = 10.44VDD1326 pKa = 3.52YY1327 pKa = 11.12HH1328 pKa = 6.78DD1329 pKa = 3.94
Molecular weight: 138.52 kDa
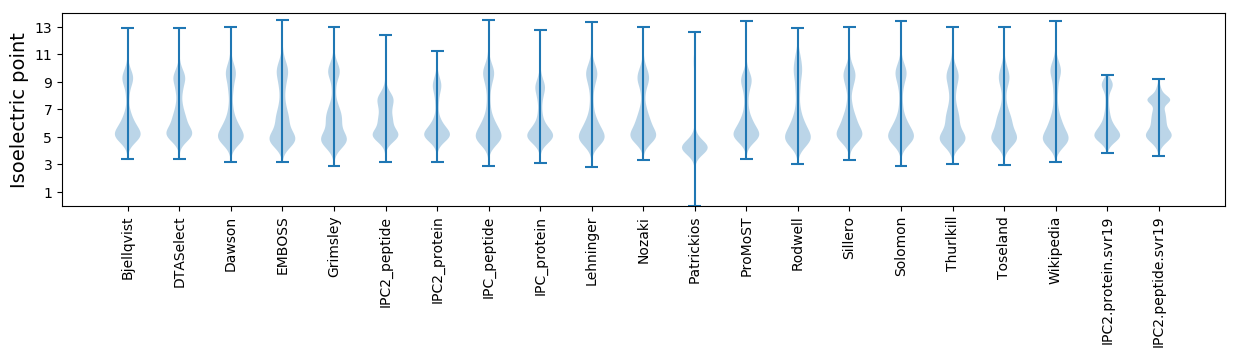
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A066ZQP2|A0A066ZQP2_HYDMR Rhodanese domain-containing protein OS=Hydrogenovibrio marinus OX=28885 GN=EI16_05825 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.49RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.76GRR39 pKa = 11.84KK40 pKa = 8.98RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.49RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.76GRR39 pKa = 11.84KK40 pKa = 8.98RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
736326 |
41 |
2103 |
316.0 |
35.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.278 ± 0.051 | 0.834 ± 0.018 |
5.602 ± 0.043 | 6.541 ± 0.052 |
4.254 ± 0.037 | 6.563 ± 0.047 |
2.302 ± 0.024 | 6.383 ± 0.04 |
6.133 ± 0.045 | 10.199 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.853 ± 0.026 | 4.237 ± 0.032 |
3.974 ± 0.029 | 4.681 ± 0.045 |
4.237 ± 0.037 | 6.416 ± 0.041 |
5.246 ± 0.039 | 6.994 ± 0.048 |
1.229 ± 0.023 | 3.044 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
