
Exophiala oligosperma
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Chaetothyriomycetidae; Chaetothyriales; Herpotrichiellaceae; Exophiala
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
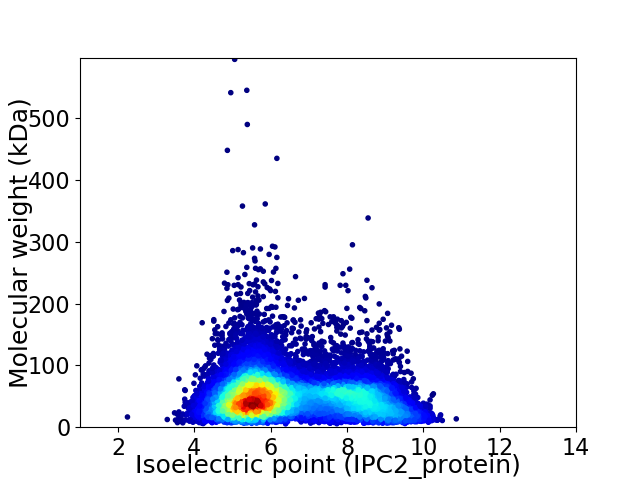
Virtual 2D-PAGE plot for 12793 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D2DRA5|A0A0D2DRA5_9EURO Uncharacterized protein OS=Exophiala oligosperma OX=215243 GN=PV06_09258 PE=4 SV=1
MM1 pKa = 7.19QRR3 pKa = 11.84TLALAALAAVAFANPMPQGVTSAIAPEE30 pKa = 4.04ASAPAGCQSTWPGTFEE46 pKa = 3.64ITVVNVTSSGKK57 pKa = 9.55RR58 pKa = 11.84DD59 pKa = 3.25LEE61 pKa = 3.96KK62 pKa = 10.55RR63 pKa = 11.84QKK65 pKa = 11.12DD66 pKa = 3.66GVLTLTLADD75 pKa = 4.54GVLKK79 pKa = 10.73DD80 pKa = 3.32QAGRR84 pKa = 11.84TGYY87 pKa = 9.86IASNYY92 pKa = 8.11QFQFDD97 pKa = 4.83DD98 pKa = 4.46PPQNGAIYY106 pKa = 9.01TKK108 pKa = 10.65GFSVCSNYY116 pKa = 10.85SLALGGDD123 pKa = 3.96ALWYY127 pKa = 9.73QCLSGSFYY135 pKa = 11.47NLYY138 pKa = 9.79NKK140 pKa = 9.74NWANTCNAIYY150 pKa = 9.67IYY152 pKa = 11.25AMSGSSAPATQAADD166 pKa = 3.98GQPQATPAPAPPVSQVSDD184 pKa = 3.81GQPQATTGVPVTQISDD200 pKa = 3.78GQPQATTGAPVSQISDD216 pKa = 3.86GQPQAPTGAPVSQISDD232 pKa = 3.86GQPQAPTGAPVSQISDD248 pKa = 3.86GQPQAPTGAPVSQISDD264 pKa = 3.86GQPQAPTGAPVSQISDD280 pKa = 3.86GQPQAPTGAPVSQISDD296 pKa = 3.86GQPQAPTGAPVSQISDD312 pKa = 3.86GQPQAPTGAPVSQISDD328 pKa = 3.86GQPQAPTGAPVSQISDD344 pKa = 3.86GQPQAPTGAPVSQISDD360 pKa = 3.86GQPQAPTGAPVSQISDD376 pKa = 3.86GQPQAPTGAPVSQISDD392 pKa = 3.86GQPQAPTGAPVSQISDD408 pKa = 3.86GQPQAPTATVEE419 pKa = 4.03AASNTGAAPATYY431 pKa = 9.07TGAANRR437 pKa = 11.84EE438 pKa = 4.17GLSGSLAVVAGLAGAMALLL457 pKa = 4.29
MM1 pKa = 7.19QRR3 pKa = 11.84TLALAALAAVAFANPMPQGVTSAIAPEE30 pKa = 4.04ASAPAGCQSTWPGTFEE46 pKa = 3.64ITVVNVTSSGKK57 pKa = 9.55RR58 pKa = 11.84DD59 pKa = 3.25LEE61 pKa = 3.96KK62 pKa = 10.55RR63 pKa = 11.84QKK65 pKa = 11.12DD66 pKa = 3.66GVLTLTLADD75 pKa = 4.54GVLKK79 pKa = 10.73DD80 pKa = 3.32QAGRR84 pKa = 11.84TGYY87 pKa = 9.86IASNYY92 pKa = 8.11QFQFDD97 pKa = 4.83DD98 pKa = 4.46PPQNGAIYY106 pKa = 9.01TKK108 pKa = 10.65GFSVCSNYY116 pKa = 10.85SLALGGDD123 pKa = 3.96ALWYY127 pKa = 9.73QCLSGSFYY135 pKa = 11.47NLYY138 pKa = 9.79NKK140 pKa = 9.74NWANTCNAIYY150 pKa = 9.67IYY152 pKa = 11.25AMSGSSAPATQAADD166 pKa = 3.98GQPQATPAPAPPVSQVSDD184 pKa = 3.81GQPQATTGVPVTQISDD200 pKa = 3.78GQPQATTGAPVSQISDD216 pKa = 3.86GQPQAPTGAPVSQISDD232 pKa = 3.86GQPQAPTGAPVSQISDD248 pKa = 3.86GQPQAPTGAPVSQISDD264 pKa = 3.86GQPQAPTGAPVSQISDD280 pKa = 3.86GQPQAPTGAPVSQISDD296 pKa = 3.86GQPQAPTGAPVSQISDD312 pKa = 3.86GQPQAPTGAPVSQISDD328 pKa = 3.86GQPQAPTGAPVSQISDD344 pKa = 3.86GQPQAPTGAPVSQISDD360 pKa = 3.86GQPQAPTGAPVSQISDD376 pKa = 3.86GQPQAPTGAPVSQISDD392 pKa = 3.86GQPQAPTGAPVSQISDD408 pKa = 3.86GQPQAPTATVEE419 pKa = 4.03AASNTGAAPATYY431 pKa = 9.07TGAANRR437 pKa = 11.84EE438 pKa = 4.17GLSGSLAVVAGLAGAMALLL457 pKa = 4.29
Molecular weight: 45.36 kDa
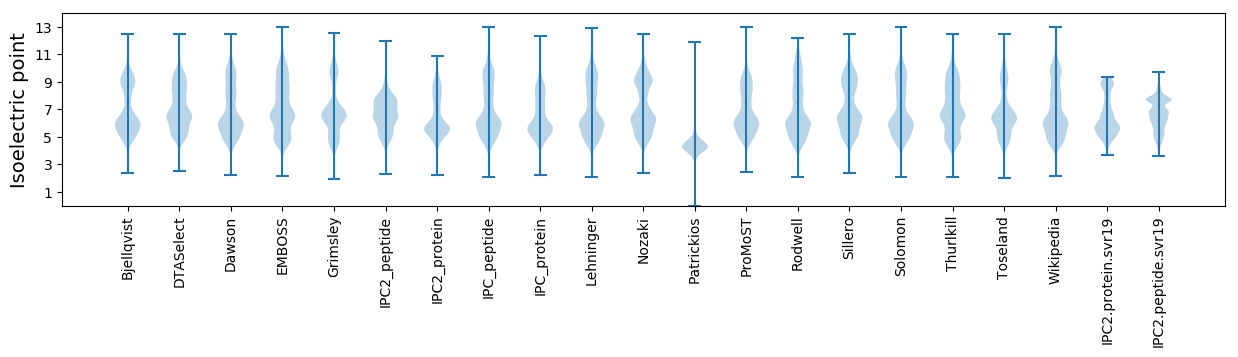
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D2DQB2|A0A0D2DQB2_9EURO Plasma membrane fusion protein PRM1 OS=Exophiala oligosperma OX=215243 GN=PV06_03431 PE=3 SV=1
MM1 pKa = 7.3VLGLGKK7 pKa = 8.57TRR9 pKa = 11.84SRR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 5.7PGGTTVTTTTTTTHH27 pKa = 6.4HH28 pKa = 6.8PAGTGAGGVARR39 pKa = 11.84KK40 pKa = 6.21PTLMQRR46 pKa = 11.84LRR48 pKa = 11.84GRR50 pKa = 11.84TTPRR54 pKa = 11.84STATTGRR61 pKa = 11.84APRR64 pKa = 11.84RR65 pKa = 11.84TRR67 pKa = 11.84RR68 pKa = 11.84TRR70 pKa = 11.84AAPATHH76 pKa = 7.03HH77 pKa = 6.19KK78 pKa = 10.05RR79 pKa = 11.84KK80 pKa = 8.33TSIGDD85 pKa = 3.67KK86 pKa = 10.84VSGAMLKK93 pKa = 10.71LKK95 pKa = 10.64GSVTHH100 pKa = 7.14RR101 pKa = 11.84PGEE104 pKa = 4.09KK105 pKa = 9.75AAGTRR110 pKa = 11.84RR111 pKa = 11.84MHH113 pKa = 5.63GTDD116 pKa = 2.88GKK118 pKa = 9.59GSRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84FYY125 pKa = 11.48
MM1 pKa = 7.3VLGLGKK7 pKa = 8.57TRR9 pKa = 11.84SRR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 5.7PGGTTVTTTTTTTHH27 pKa = 6.4HH28 pKa = 6.8PAGTGAGGVARR39 pKa = 11.84KK40 pKa = 6.21PTLMQRR46 pKa = 11.84LRR48 pKa = 11.84GRR50 pKa = 11.84TTPRR54 pKa = 11.84STATTGRR61 pKa = 11.84APRR64 pKa = 11.84RR65 pKa = 11.84TRR67 pKa = 11.84RR68 pKa = 11.84TRR70 pKa = 11.84AAPATHH76 pKa = 7.03HH77 pKa = 6.19KK78 pKa = 10.05RR79 pKa = 11.84KK80 pKa = 8.33TSIGDD85 pKa = 3.67KK86 pKa = 10.84VSGAMLKK93 pKa = 10.71LKK95 pKa = 10.64GSVTHH100 pKa = 7.14RR101 pKa = 11.84PGEE104 pKa = 4.09KK105 pKa = 9.75AAGTRR110 pKa = 11.84RR111 pKa = 11.84MHH113 pKa = 5.63GTDD116 pKa = 2.88GKK118 pKa = 9.59GSRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84FYY125 pKa = 11.48
Molecular weight: 13.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6246231 |
49 |
5476 |
488.3 |
54.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.377 ± 0.019 | 1.257 ± 0.009 |
5.782 ± 0.014 | 5.973 ± 0.02 |
3.74 ± 0.012 | 6.809 ± 0.021 |
2.479 ± 0.01 | 4.835 ± 0.014 |
4.722 ± 0.017 | 8.874 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.254 ± 0.008 | 3.656 ± 0.011 |
5.959 ± 0.021 | 4.144 ± 0.017 |
6.196 ± 0.018 | 8.392 ± 0.026 |
6.171 ± 0.016 | 6.168 ± 0.014 |
1.457 ± 0.008 | 2.753 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
