
Exophiala xenobiotica
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Chaetothyriomycetidae; Chaetothyriales; Herpotrichiellaceae; Exophiala
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
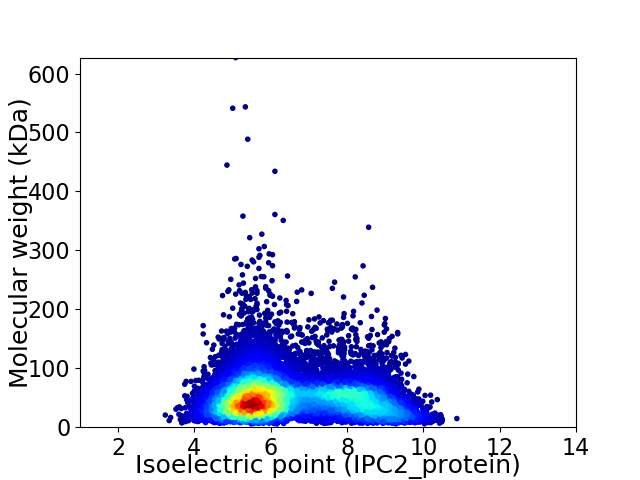
Virtual 2D-PAGE plot for 12773 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D2BHL1|A0A0D2BHL1_9EURO FAD_binding_3 domain-containing protein OS=Exophiala xenobiotica OX=348802 GN=PV05_10457 PE=4 SV=1
MM1 pKa = 7.48HH2 pKa = 7.13SLAVVLAVSALASISLAKK20 pKa = 9.64PIPVPGTSVNTKK32 pKa = 10.16KK33 pKa = 10.84GFTVNQGPAKK43 pKa = 9.51PFQPGPVQLQKK54 pKa = 11.03VYY56 pKa = 11.03SKK58 pKa = 11.23YY59 pKa = 11.08NKK61 pKa = 9.6LAPADD66 pKa = 3.75VSNAAAADD74 pKa = 4.03GSVTATPEE82 pKa = 4.18QYY84 pKa = 10.7DD85 pKa = 3.78SEE87 pKa = 4.5YY88 pKa = 10.61LSPVTIGGQTLNLDD102 pKa = 4.16FDD104 pKa = 4.24TGSADD109 pKa = 2.96LWVFSSEE116 pKa = 4.3LPSSQSSGHH125 pKa = 6.16SIYY128 pKa = 10.94NPSKK132 pKa = 10.61SSTAKK137 pKa = 10.3KK138 pKa = 10.4LSGATWDD145 pKa = 3.19ISYY148 pKa = 11.15GDD150 pKa = 3.82GSGASGDD157 pKa = 4.08VYY159 pKa = 11.04TDD161 pKa = 3.4TVDD164 pKa = 3.25VGGTTVTGQAVEE176 pKa = 3.99LAKK179 pKa = 10.23TISAQFQQDD188 pKa = 3.52QNNDD192 pKa = 3.35GLLGLAFSSINTVTPKK208 pKa = 10.25QQTTFFDD215 pKa = 3.8TAIHH219 pKa = 6.22EE220 pKa = 4.72GVLDD224 pKa = 4.16ANVFTVDD231 pKa = 4.2LKK233 pKa = 11.14KK234 pKa = 10.62GAPGSYY240 pKa = 10.59DD241 pKa = 3.18FGFIDD246 pKa = 3.9DD247 pKa = 4.01SKK249 pKa = 9.02YY250 pKa = 9.54TGEE253 pKa = 3.82ITYY256 pKa = 8.63TAVNSANGFWEE267 pKa = 4.54FTGTGYY273 pKa = 11.27GVGDD277 pKa = 4.01GSFQSSSIDD286 pKa = 3.72AIADD290 pKa = 3.48TGTTLILIDD299 pKa = 4.66DD300 pKa = 5.7DD301 pKa = 4.23IVSAYY306 pKa = 10.05YY307 pKa = 10.73DD308 pKa = 3.75EE309 pKa = 5.46VDD311 pKa = 3.14GAQYY315 pKa = 11.36DD316 pKa = 4.02NSQGGYY322 pKa = 8.2TFDD325 pKa = 4.1CTATLPDD332 pKa = 5.42FILGVEE338 pKa = 4.38DD339 pKa = 4.11AQFTVPGSFINFAPITDD356 pKa = 4.24GSSTCYY362 pKa = 10.55GGIQSNEE369 pKa = 3.64GLGLSIYY376 pKa = 11.02GDD378 pKa = 3.66VFLKK382 pKa = 10.83AVFAIFDD389 pKa = 3.74SDD391 pKa = 3.44NTQLGFANKK400 pKa = 10.13DD401 pKa = 3.29LL402 pKa = 4.31
MM1 pKa = 7.48HH2 pKa = 7.13SLAVVLAVSALASISLAKK20 pKa = 9.64PIPVPGTSVNTKK32 pKa = 10.16KK33 pKa = 10.84GFTVNQGPAKK43 pKa = 9.51PFQPGPVQLQKK54 pKa = 11.03VYY56 pKa = 11.03SKK58 pKa = 11.23YY59 pKa = 11.08NKK61 pKa = 9.6LAPADD66 pKa = 3.75VSNAAAADD74 pKa = 4.03GSVTATPEE82 pKa = 4.18QYY84 pKa = 10.7DD85 pKa = 3.78SEE87 pKa = 4.5YY88 pKa = 10.61LSPVTIGGQTLNLDD102 pKa = 4.16FDD104 pKa = 4.24TGSADD109 pKa = 2.96LWVFSSEE116 pKa = 4.3LPSSQSSGHH125 pKa = 6.16SIYY128 pKa = 10.94NPSKK132 pKa = 10.61SSTAKK137 pKa = 10.3KK138 pKa = 10.4LSGATWDD145 pKa = 3.19ISYY148 pKa = 11.15GDD150 pKa = 3.82GSGASGDD157 pKa = 4.08VYY159 pKa = 11.04TDD161 pKa = 3.4TVDD164 pKa = 3.25VGGTTVTGQAVEE176 pKa = 3.99LAKK179 pKa = 10.23TISAQFQQDD188 pKa = 3.52QNNDD192 pKa = 3.35GLLGLAFSSINTVTPKK208 pKa = 10.25QQTTFFDD215 pKa = 3.8TAIHH219 pKa = 6.22EE220 pKa = 4.72GVLDD224 pKa = 4.16ANVFTVDD231 pKa = 4.2LKK233 pKa = 11.14KK234 pKa = 10.62GAPGSYY240 pKa = 10.59DD241 pKa = 3.18FGFIDD246 pKa = 3.9DD247 pKa = 4.01SKK249 pKa = 9.02YY250 pKa = 9.54TGEE253 pKa = 3.82ITYY256 pKa = 8.63TAVNSANGFWEE267 pKa = 4.54FTGTGYY273 pKa = 11.27GVGDD277 pKa = 4.01GSFQSSSIDD286 pKa = 3.72AIADD290 pKa = 3.48TGTTLILIDD299 pKa = 4.66DD300 pKa = 5.7DD301 pKa = 4.23IVSAYY306 pKa = 10.05YY307 pKa = 10.73DD308 pKa = 3.75EE309 pKa = 5.46VDD311 pKa = 3.14GAQYY315 pKa = 11.36DD316 pKa = 4.02NSQGGYY322 pKa = 8.2TFDD325 pKa = 4.1CTATLPDD332 pKa = 5.42FILGVEE338 pKa = 4.38DD339 pKa = 4.11AQFTVPGSFINFAPITDD356 pKa = 4.24GSSTCYY362 pKa = 10.55GGIQSNEE369 pKa = 3.64GLGLSIYY376 pKa = 11.02GDD378 pKa = 3.66VFLKK382 pKa = 10.83AVFAIFDD389 pKa = 3.74SDD391 pKa = 3.44NTQLGFANKK400 pKa = 10.13DD401 pKa = 3.29LL402 pKa = 4.31
Molecular weight: 42.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D2ENP2|A0A0D2ENP2_9EURO Uncharacterized protein OS=Exophiala xenobiotica OX=348802 GN=PV05_05675 PE=4 SV=1
MM1 pKa = 7.32YY2 pKa = 9.38PPRR5 pKa = 11.84WKK7 pKa = 9.26TSPRR11 pKa = 11.84RR12 pKa = 11.84EE13 pKa = 4.03EE14 pKa = 4.05PVLCTTFTVTTTIVSTTTTIHH35 pKa = 7.28DD36 pKa = 4.51EE37 pKa = 4.03RR38 pKa = 11.84LLWPDD43 pKa = 2.84IRR45 pKa = 11.84TKK47 pKa = 10.89GKK49 pKa = 9.04WPSAQSSSSSDD60 pKa = 2.89KK61 pKa = 10.86RR62 pKa = 11.84NPRR65 pKa = 11.84LTQVGRR71 pKa = 11.84SKK73 pKa = 10.0TKK75 pKa = 10.27IRR77 pKa = 11.84WSPDD81 pKa = 2.66KK82 pKa = 10.85SSLMAFQRR90 pKa = 11.84TAMKK94 pKa = 10.83AEE96 pKa = 4.05TQRR99 pKa = 11.84PKK101 pKa = 10.89SLEE104 pKa = 3.88TTTCSRR110 pKa = 11.84NQKK113 pKa = 7.88TCYY116 pKa = 9.75LCCFLHH122 pKa = 7.68RR123 pKa = 11.84IRR125 pKa = 11.84PHH127 pKa = 6.04YY128 pKa = 9.59GRR130 pKa = 11.84KK131 pKa = 9.29SDD133 pKa = 4.69LGRR136 pKa = 11.84SGQGVCHH143 pKa = 6.05EE144 pKa = 4.53EE145 pKa = 3.88KK146 pKa = 10.85VYY148 pKa = 10.79APSLINVYY156 pKa = 9.44TNLSIFVLFCVRR168 pKa = 11.84SSIINLANTPLALGHH183 pKa = 6.48RR184 pKa = 11.84CRR186 pKa = 11.84SRR188 pKa = 11.84QTFRR192 pKa = 11.84PLRR195 pKa = 11.84VPRR198 pKa = 11.84MGVVGVLRR206 pKa = 11.84RR207 pKa = 11.84PPCSPPPPPPPPVGCPPRR225 pKa = 11.84TPQPLVVQASDD236 pKa = 3.39HH237 pKa = 6.13RR238 pKa = 11.84CRR240 pKa = 11.84PASGHH245 pKa = 6.06SVTTFNTSMASTMPLRR261 pKa = 11.84CSEE264 pKa = 4.24LL265 pKa = 3.47
MM1 pKa = 7.32YY2 pKa = 9.38PPRR5 pKa = 11.84WKK7 pKa = 9.26TSPRR11 pKa = 11.84RR12 pKa = 11.84EE13 pKa = 4.03EE14 pKa = 4.05PVLCTTFTVTTTIVSTTTTIHH35 pKa = 7.28DD36 pKa = 4.51EE37 pKa = 4.03RR38 pKa = 11.84LLWPDD43 pKa = 2.84IRR45 pKa = 11.84TKK47 pKa = 10.89GKK49 pKa = 9.04WPSAQSSSSSDD60 pKa = 2.89KK61 pKa = 10.86RR62 pKa = 11.84NPRR65 pKa = 11.84LTQVGRR71 pKa = 11.84SKK73 pKa = 10.0TKK75 pKa = 10.27IRR77 pKa = 11.84WSPDD81 pKa = 2.66KK82 pKa = 10.85SSLMAFQRR90 pKa = 11.84TAMKK94 pKa = 10.83AEE96 pKa = 4.05TQRR99 pKa = 11.84PKK101 pKa = 10.89SLEE104 pKa = 3.88TTTCSRR110 pKa = 11.84NQKK113 pKa = 7.88TCYY116 pKa = 9.75LCCFLHH122 pKa = 7.68RR123 pKa = 11.84IRR125 pKa = 11.84PHH127 pKa = 6.04YY128 pKa = 9.59GRR130 pKa = 11.84KK131 pKa = 9.29SDD133 pKa = 4.69LGRR136 pKa = 11.84SGQGVCHH143 pKa = 6.05EE144 pKa = 4.53EE145 pKa = 3.88KK146 pKa = 10.85VYY148 pKa = 10.79APSLINVYY156 pKa = 9.44TNLSIFVLFCVRR168 pKa = 11.84SSIINLANTPLALGHH183 pKa = 6.48RR184 pKa = 11.84CRR186 pKa = 11.84SRR188 pKa = 11.84QTFRR192 pKa = 11.84PLRR195 pKa = 11.84VPRR198 pKa = 11.84MGVVGVLRR206 pKa = 11.84RR207 pKa = 11.84PPCSPPPPPPPPVGCPPRR225 pKa = 11.84TPQPLVVQASDD236 pKa = 3.39HH237 pKa = 6.13RR238 pKa = 11.84CRR240 pKa = 11.84PASGHH245 pKa = 6.06SVTTFNTSMASTMPLRR261 pKa = 11.84CSEE264 pKa = 4.24LL265 pKa = 3.47
Molecular weight: 29.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6285281 |
49 |
5762 |
492.1 |
54.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.602 ± 0.016 | 1.246 ± 0.008 |
5.701 ± 0.014 | 6.048 ± 0.019 |
3.726 ± 0.013 | 6.83 ± 0.019 |
2.465 ± 0.008 | 4.837 ± 0.013 |
4.791 ± 0.02 | 8.876 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.26 ± 0.008 | 3.599 ± 0.011 |
6.03 ± 0.021 | 4.166 ± 0.015 |
6.159 ± 0.018 | 8.214 ± 0.026 |
6.062 ± 0.015 | 6.121 ± 0.013 |
1.477 ± 0.007 | 2.789 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
