
Hubei sobemo-like virus 48
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.48
Get precalculated fractions of proteins
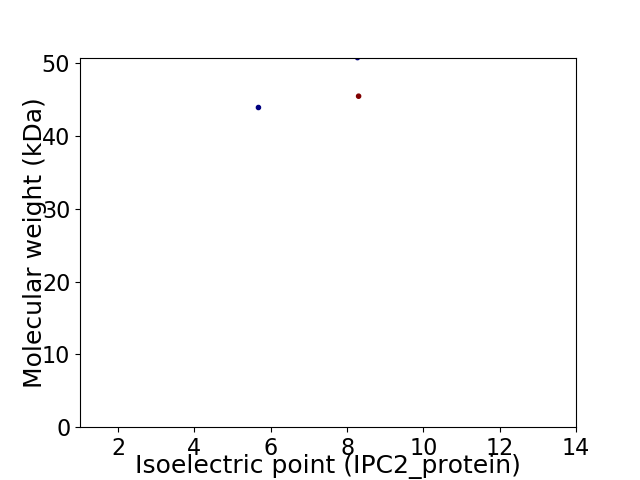
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
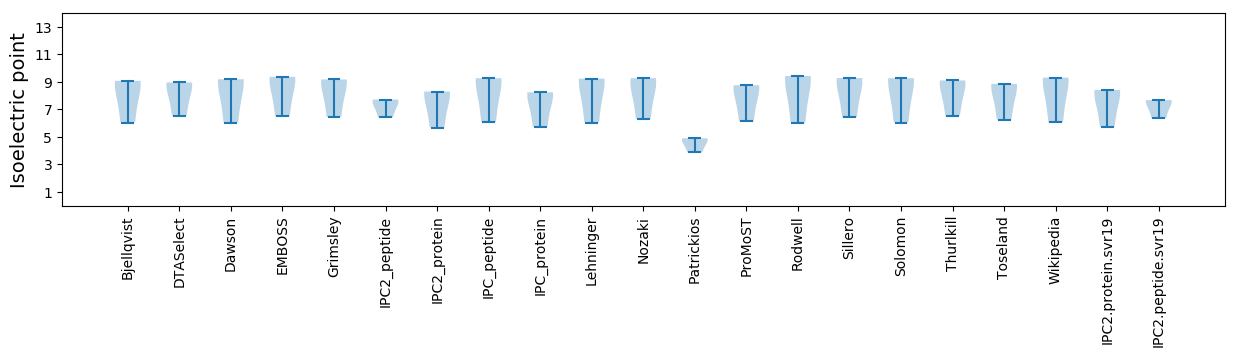
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEH0|A0A1L3KEH0_9VIRU Capsid protein OS=Hubei sobemo-like virus 48 OX=1923236 PE=3 SV=1
MM1 pKa = 6.84EE2 pKa = 3.96QAYY5 pKa = 10.6SPVVWSLPDD14 pKa = 4.27DD15 pKa = 4.23FDD17 pKa = 4.55SRR19 pKa = 11.84TRR21 pKa = 11.84FDD23 pKa = 3.01WAIRR27 pKa = 11.84RR28 pKa = 11.84LDD30 pKa = 3.7MQSSPGMPYY39 pKa = 9.35MRR41 pKa = 11.84EE42 pKa = 3.79APTNGKK48 pKa = 6.63WLKK51 pKa = 9.55WDD53 pKa = 3.39GVEE56 pKa = 4.11YY57 pKa = 10.93DD58 pKa = 3.65AMQANRR64 pKa = 11.84LWHH67 pKa = 7.05DD68 pKa = 3.87VQCVLSDD75 pKa = 3.42DD76 pKa = 3.94WEE78 pKa = 4.15HH79 pKa = 6.8VIRR82 pKa = 11.84VFIKK86 pKa = 9.76QEE88 pKa = 3.49PHH90 pKa = 6.4KK91 pKa = 10.22KK92 pKa = 9.3HH93 pKa = 6.39KK94 pKa = 10.47ARR96 pKa = 11.84EE97 pKa = 4.26GRR99 pKa = 11.84WRR101 pKa = 11.84LIMASSLPVQLVWHH115 pKa = 6.0MLFSYY120 pKa = 10.53MNDD123 pKa = 3.45LEE125 pKa = 4.98ISEE128 pKa = 5.41CYY130 pKa = 10.13NIPSQHH136 pKa = 5.71GLILVGGGWKK146 pKa = 10.2DD147 pKa = 3.61YY148 pKa = 10.58LRR150 pKa = 11.84SWKK153 pKa = 10.54EE154 pKa = 3.46EE155 pKa = 3.91GLSVGLDD162 pKa = 3.12KK163 pKa = 11.37SAWDD167 pKa = 3.23WTAPRR172 pKa = 11.84WVMDD176 pKa = 3.14WDD178 pKa = 3.86LDD180 pKa = 3.39FRR182 pKa = 11.84YY183 pKa = 10.57RR184 pKa = 11.84MGRR187 pKa = 11.84GKK189 pKa = 10.61RR190 pKa = 11.84MEE192 pKa = 3.93EE193 pKa = 3.4WHH195 pKa = 6.81RR196 pKa = 11.84LAKK199 pKa = 10.5LMYY202 pKa = 9.63HH203 pKa = 6.78HH204 pKa = 6.74MFDD207 pKa = 4.26HH208 pKa = 7.22PVLQLSDD215 pKa = 3.59GTLLRR220 pKa = 11.84QTVPGIMKK228 pKa = 9.74SGCVNTISTNGHH240 pKa = 5.42AQVMMHH246 pKa = 6.29CVVAEE251 pKa = 4.2DD252 pKa = 4.03SNVPYY257 pKa = 10.44EE258 pKa = 4.87PYY260 pKa = 10.42PKK262 pKa = 9.74TCGDD266 pKa = 3.85DD267 pKa = 3.7TLEE270 pKa = 5.04HH271 pKa = 6.64PMHH274 pKa = 5.85TQSLEE279 pKa = 3.66YY280 pKa = 9.01YY281 pKa = 8.33GRR283 pKa = 11.84YY284 pKa = 8.76GVVVKK289 pKa = 10.63SVSEE293 pKa = 3.71NMEE296 pKa = 4.12FVGHH300 pKa = 6.8EE301 pKa = 4.17FTDD304 pKa = 3.94SGPHH308 pKa = 6.24PLYY311 pKa = 10.24ISKK314 pKa = 10.18HH315 pKa = 4.23MKK317 pKa = 9.49KK318 pKa = 9.95LQYY321 pKa = 10.49LADD324 pKa = 5.27DD325 pKa = 5.62IIPDD329 pKa = 4.12FLDD332 pKa = 3.18AMARR336 pKa = 11.84MYY338 pKa = 11.09VHH340 pKa = 6.62TRR342 pKa = 11.84YY343 pKa = 10.01FSIWEE348 pKa = 4.02DD349 pKa = 3.29LAIVNGTPLPMSKK362 pKa = 8.88MAYY365 pKa = 9.42RR366 pKa = 11.84YY367 pKa = 9.17WYY369 pKa = 9.74DD370 pKa = 3.4FSVV373 pKa = 3.05
MM1 pKa = 6.84EE2 pKa = 3.96QAYY5 pKa = 10.6SPVVWSLPDD14 pKa = 4.27DD15 pKa = 4.23FDD17 pKa = 4.55SRR19 pKa = 11.84TRR21 pKa = 11.84FDD23 pKa = 3.01WAIRR27 pKa = 11.84RR28 pKa = 11.84LDD30 pKa = 3.7MQSSPGMPYY39 pKa = 9.35MRR41 pKa = 11.84EE42 pKa = 3.79APTNGKK48 pKa = 6.63WLKK51 pKa = 9.55WDD53 pKa = 3.39GVEE56 pKa = 4.11YY57 pKa = 10.93DD58 pKa = 3.65AMQANRR64 pKa = 11.84LWHH67 pKa = 7.05DD68 pKa = 3.87VQCVLSDD75 pKa = 3.42DD76 pKa = 3.94WEE78 pKa = 4.15HH79 pKa = 6.8VIRR82 pKa = 11.84VFIKK86 pKa = 9.76QEE88 pKa = 3.49PHH90 pKa = 6.4KK91 pKa = 10.22KK92 pKa = 9.3HH93 pKa = 6.39KK94 pKa = 10.47ARR96 pKa = 11.84EE97 pKa = 4.26GRR99 pKa = 11.84WRR101 pKa = 11.84LIMASSLPVQLVWHH115 pKa = 6.0MLFSYY120 pKa = 10.53MNDD123 pKa = 3.45LEE125 pKa = 4.98ISEE128 pKa = 5.41CYY130 pKa = 10.13NIPSQHH136 pKa = 5.71GLILVGGGWKK146 pKa = 10.2DD147 pKa = 3.61YY148 pKa = 10.58LRR150 pKa = 11.84SWKK153 pKa = 10.54EE154 pKa = 3.46EE155 pKa = 3.91GLSVGLDD162 pKa = 3.12KK163 pKa = 11.37SAWDD167 pKa = 3.23WTAPRR172 pKa = 11.84WVMDD176 pKa = 3.14WDD178 pKa = 3.86LDD180 pKa = 3.39FRR182 pKa = 11.84YY183 pKa = 10.57RR184 pKa = 11.84MGRR187 pKa = 11.84GKK189 pKa = 10.61RR190 pKa = 11.84MEE192 pKa = 3.93EE193 pKa = 3.4WHH195 pKa = 6.81RR196 pKa = 11.84LAKK199 pKa = 10.5LMYY202 pKa = 9.63HH203 pKa = 6.78HH204 pKa = 6.74MFDD207 pKa = 4.26HH208 pKa = 7.22PVLQLSDD215 pKa = 3.59GTLLRR220 pKa = 11.84QTVPGIMKK228 pKa = 9.74SGCVNTISTNGHH240 pKa = 5.42AQVMMHH246 pKa = 6.29CVVAEE251 pKa = 4.2DD252 pKa = 4.03SNVPYY257 pKa = 10.44EE258 pKa = 4.87PYY260 pKa = 10.42PKK262 pKa = 9.74TCGDD266 pKa = 3.85DD267 pKa = 3.7TLEE270 pKa = 5.04HH271 pKa = 6.64PMHH274 pKa = 5.85TQSLEE279 pKa = 3.66YY280 pKa = 9.01YY281 pKa = 8.33GRR283 pKa = 11.84YY284 pKa = 8.76GVVVKK289 pKa = 10.63SVSEE293 pKa = 3.71NMEE296 pKa = 4.12FVGHH300 pKa = 6.8EE301 pKa = 4.17FTDD304 pKa = 3.94SGPHH308 pKa = 6.24PLYY311 pKa = 10.24ISKK314 pKa = 10.18HH315 pKa = 4.23MKK317 pKa = 9.49KK318 pKa = 9.95LQYY321 pKa = 10.49LADD324 pKa = 5.27DD325 pKa = 5.62IIPDD329 pKa = 4.12FLDD332 pKa = 3.18AMARR336 pKa = 11.84MYY338 pKa = 11.09VHH340 pKa = 6.62TRR342 pKa = 11.84YY343 pKa = 10.01FSIWEE348 pKa = 4.02DD349 pKa = 3.29LAIVNGTPLPMSKK362 pKa = 8.88MAYY365 pKa = 9.42RR366 pKa = 11.84YY367 pKa = 9.17WYY369 pKa = 9.74DD370 pKa = 3.4FSVV373 pKa = 3.05
Molecular weight: 43.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEG3|A0A1L3KEG3_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 48 OX=1923236 PE=4 SV=1
MM1 pKa = 7.19SWKK4 pKa = 10.45DD5 pKa = 3.5YY6 pKa = 10.95LLVSGGVYY14 pKa = 9.88GLWMGHH20 pKa = 6.39KK21 pKa = 9.6IGVGSKK27 pKa = 8.42ISAVAGMVIPGFRR40 pKa = 11.84KK41 pKa = 10.13LKK43 pKa = 8.03WAVGRR48 pKa = 11.84AEE50 pKa = 4.64VVSQPAASNHH60 pKa = 5.44LRR62 pKa = 11.84LEE64 pKa = 4.02SRR66 pKa = 11.84KK67 pKa = 10.44DD68 pKa = 3.32GSEE71 pKa = 4.06EE72 pKa = 3.96VSLTAPRR79 pKa = 11.84YY80 pKa = 8.94QVIVCEE86 pKa = 4.39KK87 pKa = 10.17KK88 pKa = 10.32DD89 pKa = 3.54GRR91 pKa = 11.84LVKK94 pKa = 10.44LGCAVRR100 pKa = 11.84FDD102 pKa = 4.3GNFLVGPDD110 pKa = 3.86HH111 pKa = 7.14VLGEE115 pKa = 4.36ADD117 pKa = 3.61LPKK120 pKa = 10.61YY121 pKa = 10.57AVGRR125 pKa = 11.84QKK127 pKa = 10.27FVCLDD132 pKa = 3.22KK133 pKa = 11.22KK134 pKa = 10.87EE135 pKa = 4.71RR136 pKa = 11.84IPLDD140 pKa = 3.36TDD142 pKa = 3.92LVAIRR147 pKa = 11.84MSPSEE152 pKa = 4.14LSTIGVSVAKK162 pKa = 10.25IGLVSAKK169 pKa = 9.97GVYY172 pKa = 9.18SQIVGVDD179 pKa = 3.45SKK181 pKa = 11.32GTTGVLKK188 pKa = 9.71MDD190 pKa = 3.9RR191 pKa = 11.84VAFGRR196 pKa = 11.84TVYY199 pKa = 10.82DD200 pKa = 3.52GTTLPGYY207 pKa = 10.27SGAAYY212 pKa = 9.29TSGAFCSAIHH222 pKa = 5.63QSGGAVNGGYY232 pKa = 10.31ASSYY236 pKa = 9.79IWMLLKK242 pKa = 10.83DD243 pKa = 3.96HH244 pKa = 6.84LRR246 pKa = 11.84VEE248 pKa = 4.74DD249 pKa = 3.8VVEE252 pKa = 4.28PEE254 pKa = 5.09SKK256 pKa = 10.84KK257 pKa = 10.5NWNSDD262 pKa = 3.23TPEE265 pKa = 3.74WLLSQFKK272 pKa = 10.48AGKK275 pKa = 7.92KK276 pKa = 9.66LKK278 pKa = 9.24WKK280 pKa = 9.21RR281 pKa = 11.84TGDD284 pKa = 3.57PDD286 pKa = 5.6LIEE289 pKa = 5.74LMLDD293 pKa = 3.23DD294 pKa = 5.03GKK296 pKa = 11.0FSRR299 pKa = 11.84VSPASMHH306 pKa = 6.43KK307 pKa = 10.68AFGPTWHH314 pKa = 7.13DD315 pKa = 2.78HH316 pKa = 6.63DD317 pKa = 5.17VIEE320 pKa = 4.79KK321 pKa = 10.64GFDD324 pKa = 2.69RR325 pKa = 11.84SYY327 pKa = 11.05RR328 pKa = 11.84DD329 pKa = 3.33VPRR332 pKa = 11.84EE333 pKa = 4.0SILEE337 pKa = 4.12SVPCTSGSGEE347 pKa = 4.01DD348 pKa = 4.25HH349 pKa = 6.79GSKK352 pKa = 10.81LPGALSLLEE361 pKa = 4.91PDD363 pKa = 3.77QAWARR368 pKa = 11.84LSLQQEE374 pKa = 3.73MRR376 pKa = 11.84EE377 pKa = 3.9FFNLSKK383 pKa = 10.55RR384 pKa = 11.84QQEE387 pKa = 4.57DD388 pKa = 2.7IRR390 pKa = 11.84KK391 pKa = 8.69CSQRR395 pKa = 11.84QMSQNLASSGQAKK408 pKa = 8.86TMATKK413 pKa = 10.77NSS415 pKa = 3.54
MM1 pKa = 7.19SWKK4 pKa = 10.45DD5 pKa = 3.5YY6 pKa = 10.95LLVSGGVYY14 pKa = 9.88GLWMGHH20 pKa = 6.39KK21 pKa = 9.6IGVGSKK27 pKa = 8.42ISAVAGMVIPGFRR40 pKa = 11.84KK41 pKa = 10.13LKK43 pKa = 8.03WAVGRR48 pKa = 11.84AEE50 pKa = 4.64VVSQPAASNHH60 pKa = 5.44LRR62 pKa = 11.84LEE64 pKa = 4.02SRR66 pKa = 11.84KK67 pKa = 10.44DD68 pKa = 3.32GSEE71 pKa = 4.06EE72 pKa = 3.96VSLTAPRR79 pKa = 11.84YY80 pKa = 8.94QVIVCEE86 pKa = 4.39KK87 pKa = 10.17KK88 pKa = 10.32DD89 pKa = 3.54GRR91 pKa = 11.84LVKK94 pKa = 10.44LGCAVRR100 pKa = 11.84FDD102 pKa = 4.3GNFLVGPDD110 pKa = 3.86HH111 pKa = 7.14VLGEE115 pKa = 4.36ADD117 pKa = 3.61LPKK120 pKa = 10.61YY121 pKa = 10.57AVGRR125 pKa = 11.84QKK127 pKa = 10.27FVCLDD132 pKa = 3.22KK133 pKa = 11.22KK134 pKa = 10.87EE135 pKa = 4.71RR136 pKa = 11.84IPLDD140 pKa = 3.36TDD142 pKa = 3.92LVAIRR147 pKa = 11.84MSPSEE152 pKa = 4.14LSTIGVSVAKK162 pKa = 10.25IGLVSAKK169 pKa = 9.97GVYY172 pKa = 9.18SQIVGVDD179 pKa = 3.45SKK181 pKa = 11.32GTTGVLKK188 pKa = 9.71MDD190 pKa = 3.9RR191 pKa = 11.84VAFGRR196 pKa = 11.84TVYY199 pKa = 10.82DD200 pKa = 3.52GTTLPGYY207 pKa = 10.27SGAAYY212 pKa = 9.29TSGAFCSAIHH222 pKa = 5.63QSGGAVNGGYY232 pKa = 10.31ASSYY236 pKa = 9.79IWMLLKK242 pKa = 10.83DD243 pKa = 3.96HH244 pKa = 6.84LRR246 pKa = 11.84VEE248 pKa = 4.74DD249 pKa = 3.8VVEE252 pKa = 4.28PEE254 pKa = 5.09SKK256 pKa = 10.84KK257 pKa = 10.5NWNSDD262 pKa = 3.23TPEE265 pKa = 3.74WLLSQFKK272 pKa = 10.48AGKK275 pKa = 7.92KK276 pKa = 9.66LKK278 pKa = 9.24WKK280 pKa = 9.21RR281 pKa = 11.84TGDD284 pKa = 3.57PDD286 pKa = 5.6LIEE289 pKa = 5.74LMLDD293 pKa = 3.23DD294 pKa = 5.03GKK296 pKa = 11.0FSRR299 pKa = 11.84VSPASMHH306 pKa = 6.43KK307 pKa = 10.68AFGPTWHH314 pKa = 7.13DD315 pKa = 2.78HH316 pKa = 6.63DD317 pKa = 5.17VIEE320 pKa = 4.79KK321 pKa = 10.64GFDD324 pKa = 2.69RR325 pKa = 11.84SYY327 pKa = 11.05RR328 pKa = 11.84DD329 pKa = 3.33VPRR332 pKa = 11.84EE333 pKa = 4.0SILEE337 pKa = 4.12SVPCTSGSGEE347 pKa = 4.01DD348 pKa = 4.25HH349 pKa = 6.79GSKK352 pKa = 10.81LPGALSLLEE361 pKa = 4.91PDD363 pKa = 3.77QAWARR368 pKa = 11.84LSLQQEE374 pKa = 3.73MRR376 pKa = 11.84EE377 pKa = 3.9FFNLSKK383 pKa = 10.55RR384 pKa = 11.84QQEE387 pKa = 4.57DD388 pKa = 2.7IRR390 pKa = 11.84KK391 pKa = 8.69CSQRR395 pKa = 11.84QMSQNLASSGQAKK408 pKa = 8.86TMATKK413 pKa = 10.77NSS415 pKa = 3.54
Molecular weight: 45.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1252 |
373 |
464 |
417.3 |
46.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.188 ± 1.037 | 1.038 ± 0.24 |
6.31 ± 0.55 | 5.032 ± 0.131 |
3.355 ± 0.292 | 7.907 ± 0.86 |
2.636 ± 0.773 | 4.313 ± 0.425 |
5.99 ± 0.751 | 8.626 ± 0.204 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.355 ± 0.985 | 2.316 ± 0.15 |
5.112 ± 0.392 | 3.914 ± 0.336 |
5.751 ± 0.082 | 8.706 ± 0.68 |
3.994 ± 0.295 | 7.987 ± 0.268 |
2.875 ± 0.584 | 3.514 ± 0.644 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
