
Fonsecaea erecta
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Chaetothyriomycetidae; Chaetothyriales; Herpotrichiellaceae; Fonsecaea
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
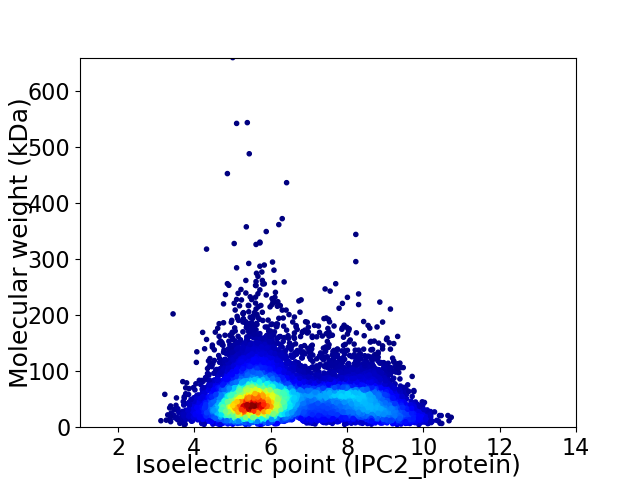
Virtual 2D-PAGE plot for 12089 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A178ZQF3|A0A178ZQF3_9EURO ULP_PROTEASE domain-containing protein OS=Fonsecaea erecta OX=1367422 GN=AYL99_03894 PE=3 SV=1
MM1 pKa = 7.83IPILLCSLLLALVQFGVSQTVSSSDD26 pKa = 3.13GYY28 pKa = 9.14TGYY31 pKa = 10.9SLSVTGNDD39 pKa = 3.02TDD41 pKa = 3.83SAVYY45 pKa = 8.31EE46 pKa = 4.29TEE48 pKa = 4.03EE49 pKa = 4.35TDD51 pKa = 3.44TSDD54 pKa = 4.6GAVTDD59 pKa = 4.24ASTPPDD65 pKa = 3.45VYY67 pKa = 11.34LNASVSVGEE76 pKa = 4.38IDD78 pKa = 3.22ITVKK82 pKa = 10.74NLSAKK87 pKa = 10.22INIDD91 pKa = 3.52AQVLSLLQFNAGVDD105 pKa = 3.6LSINAVVLNIQNVTAKK121 pKa = 10.59VEE123 pKa = 4.04LEE125 pKa = 3.71ARR127 pKa = 11.84LEE129 pKa = 3.92NLVLMINDD137 pKa = 3.94TLNSIDD143 pKa = 4.87LNPIIATLGSDD154 pKa = 3.21VGSLVGTALGGSSTSSSSASSSTSGLSARR183 pKa = 11.84SVLLDD188 pKa = 3.3EE189 pKa = 5.7GILYY193 pKa = 10.36SVNDD197 pKa = 3.67YY198 pKa = 11.33SGNTHH203 pKa = 6.2TNRR206 pKa = 11.84ILDD209 pKa = 3.53QAGNIVDD216 pKa = 3.9QKK218 pKa = 10.56LHH220 pKa = 6.63NDD222 pKa = 3.03GSVYY226 pKa = 10.36SSQVVGSYY234 pKa = 6.62WHH236 pKa = 7.24DD237 pKa = 3.32MAFTGHH243 pKa = 5.46EE244 pKa = 3.91RR245 pKa = 11.84AVLFNGEE252 pKa = 3.97NAKK255 pKa = 10.11EE256 pKa = 3.99LEE258 pKa = 4.23YY259 pKa = 10.8SYY261 pKa = 11.78SPFNGFSSIAAIYY274 pKa = 10.28QDD276 pKa = 3.08AQGNVLGTRR285 pKa = 11.84VLSEE289 pKa = 3.94IEE291 pKa = 4.21GGGSSTIADD300 pKa = 3.81DD301 pKa = 3.65
MM1 pKa = 7.83IPILLCSLLLALVQFGVSQTVSSSDD26 pKa = 3.13GYY28 pKa = 9.14TGYY31 pKa = 10.9SLSVTGNDD39 pKa = 3.02TDD41 pKa = 3.83SAVYY45 pKa = 8.31EE46 pKa = 4.29TEE48 pKa = 4.03EE49 pKa = 4.35TDD51 pKa = 3.44TSDD54 pKa = 4.6GAVTDD59 pKa = 4.24ASTPPDD65 pKa = 3.45VYY67 pKa = 11.34LNASVSVGEE76 pKa = 4.38IDD78 pKa = 3.22ITVKK82 pKa = 10.74NLSAKK87 pKa = 10.22INIDD91 pKa = 3.52AQVLSLLQFNAGVDD105 pKa = 3.6LSINAVVLNIQNVTAKK121 pKa = 10.59VEE123 pKa = 4.04LEE125 pKa = 3.71ARR127 pKa = 11.84LEE129 pKa = 3.92NLVLMINDD137 pKa = 3.94TLNSIDD143 pKa = 4.87LNPIIATLGSDD154 pKa = 3.21VGSLVGTALGGSSTSSSSASSSTSGLSARR183 pKa = 11.84SVLLDD188 pKa = 3.3EE189 pKa = 5.7GILYY193 pKa = 10.36SVNDD197 pKa = 3.67YY198 pKa = 11.33SGNTHH203 pKa = 6.2TNRR206 pKa = 11.84ILDD209 pKa = 3.53QAGNIVDD216 pKa = 3.9QKK218 pKa = 10.56LHH220 pKa = 6.63NDD222 pKa = 3.03GSVYY226 pKa = 10.36SSQVVGSYY234 pKa = 6.62WHH236 pKa = 7.24DD237 pKa = 3.32MAFTGHH243 pKa = 5.46EE244 pKa = 3.91RR245 pKa = 11.84AVLFNGEE252 pKa = 3.97NAKK255 pKa = 10.11EE256 pKa = 3.99LEE258 pKa = 4.23YY259 pKa = 10.8SYY261 pKa = 11.78SPFNGFSSIAAIYY274 pKa = 10.28QDD276 pKa = 3.08AQGNVLGTRR285 pKa = 11.84VLSEE289 pKa = 3.94IEE291 pKa = 4.21GGGSSTIADD300 pKa = 3.81DD301 pKa = 3.65
Molecular weight: 31.5 kDa
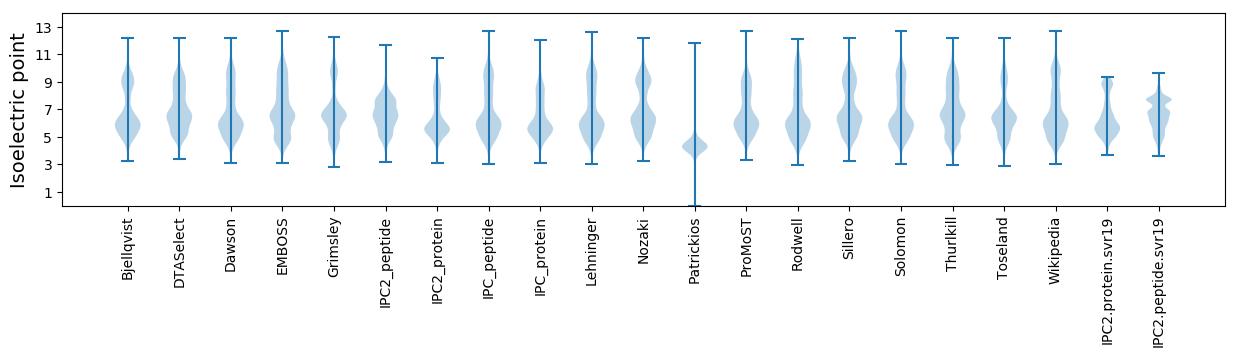
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A178Z9G9|A0A178Z9G9_9EURO NodB homology domain-containing protein OS=Fonsecaea erecta OX=1367422 GN=AYL99_09608 PE=4 SV=1
MM1 pKa = 7.15HH2 pKa = 7.78RR3 pKa = 11.84EE4 pKa = 3.78THH6 pKa = 5.49IHH8 pKa = 6.86LKK10 pKa = 7.56TLKK13 pKa = 9.86PLLRR17 pKa = 11.84AYY19 pKa = 10.41ALGYY23 pKa = 8.8LTVTAPRR30 pKa = 11.84LFGFLRR36 pKa = 11.84TLRR39 pKa = 11.84RR40 pKa = 11.84SHH42 pKa = 6.12LASRR46 pKa = 11.84EE47 pKa = 3.93VFSILRR53 pKa = 11.84RR54 pKa = 11.84ILITSTQLNRR64 pKa = 11.84FPTAAAVIVGGATALPRR81 pKa = 11.84LLLATLEE88 pKa = 4.53GVLSLLRR95 pKa = 11.84KK96 pKa = 9.88GPPTRR101 pKa = 11.84LSRR104 pKa = 11.84SIVGLIHH111 pKa = 6.34FLCSFWSAWLAFALLNRR128 pKa = 11.84DD129 pKa = 3.62EE130 pKa = 4.21TWVRR134 pKa = 11.84KK135 pKa = 9.2RR136 pKa = 11.84AISRR140 pKa = 11.84ASPNTDD146 pKa = 2.51ASALGASTLHH156 pKa = 5.83QPSPPSYY163 pKa = 8.57HH164 pKa = 6.21PHH166 pKa = 5.92YY167 pKa = 10.34AGKK170 pKa = 8.75TIDD173 pKa = 3.48FTAFAFCRR181 pKa = 11.84AVDD184 pKa = 3.71VMVITLWTRR193 pKa = 11.84TRR195 pKa = 11.84TRR197 pKa = 11.84SWHH200 pKa = 5.7PEE202 pKa = 3.32QRR204 pKa = 11.84NPRR207 pKa = 11.84LSNLVRR213 pKa = 11.84KK214 pKa = 9.02LADD217 pKa = 3.54PSVFAGSAAIIMWSWFYY234 pKa = 11.36SPEE237 pKa = 3.93RR238 pKa = 11.84LPRR241 pKa = 11.84SYY243 pKa = 11.04NQWISKK249 pKa = 10.07AADD252 pKa = 3.12IDD254 pKa = 3.64PRR256 pKa = 11.84LIQALRR262 pKa = 11.84LARR265 pKa = 11.84QGNFVYY271 pKa = 11.02GEE273 pKa = 4.2DD274 pKa = 3.47TGQAPLLAGLCRR286 pKa = 11.84EE287 pKa = 4.19LDD289 pKa = 3.8LPEE292 pKa = 4.69EE293 pKa = 4.33FADD296 pKa = 4.0PSKK299 pKa = 9.97TIPIPCEE306 pKa = 3.86LYY308 pKa = 9.99HH309 pKa = 6.54CGSGKK314 pKa = 9.94SCEE317 pKa = 3.69IHH319 pKa = 6.09ALSRR323 pKa = 11.84FWRR326 pKa = 11.84SWKK329 pKa = 9.96FAMEE333 pKa = 4.26IYY335 pKa = 10.71LPLQLLTILRR345 pKa = 11.84SPRR348 pKa = 11.84PKK350 pKa = 10.44SVLKK354 pKa = 10.54VLQAASRR361 pKa = 11.84SSSFLSGFVASFYY374 pKa = 10.97YY375 pKa = 10.16AVCLARR381 pKa = 11.84TRR383 pKa = 11.84LGPKK387 pKa = 9.43IFSQRR392 pKa = 11.84TVTPQMWDD400 pKa = 2.58SGLCVLTGCLACGWSVLLEE419 pKa = 4.3KK420 pKa = 10.34PSRR423 pKa = 11.84RR424 pKa = 11.84QEE426 pKa = 3.51IAFFVAPRR434 pKa = 11.84ALATILPRR442 pKa = 11.84VYY444 pKa = 10.06DD445 pKa = 3.07QQYY448 pKa = 9.05RR449 pKa = 11.84HH450 pKa = 6.41RR451 pKa = 11.84EE452 pKa = 3.65QAIFATSIAVVLTALKK468 pKa = 10.53SGNEE472 pKa = 3.73RR473 pKa = 11.84NIRR476 pKa = 11.84GVLGRR481 pKa = 11.84ILGGILKK488 pKa = 10.22DD489 pKa = 3.5
MM1 pKa = 7.15HH2 pKa = 7.78RR3 pKa = 11.84EE4 pKa = 3.78THH6 pKa = 5.49IHH8 pKa = 6.86LKK10 pKa = 7.56TLKK13 pKa = 9.86PLLRR17 pKa = 11.84AYY19 pKa = 10.41ALGYY23 pKa = 8.8LTVTAPRR30 pKa = 11.84LFGFLRR36 pKa = 11.84TLRR39 pKa = 11.84RR40 pKa = 11.84SHH42 pKa = 6.12LASRR46 pKa = 11.84EE47 pKa = 3.93VFSILRR53 pKa = 11.84RR54 pKa = 11.84ILITSTQLNRR64 pKa = 11.84FPTAAAVIVGGATALPRR81 pKa = 11.84LLLATLEE88 pKa = 4.53GVLSLLRR95 pKa = 11.84KK96 pKa = 9.88GPPTRR101 pKa = 11.84LSRR104 pKa = 11.84SIVGLIHH111 pKa = 6.34FLCSFWSAWLAFALLNRR128 pKa = 11.84DD129 pKa = 3.62EE130 pKa = 4.21TWVRR134 pKa = 11.84KK135 pKa = 9.2RR136 pKa = 11.84AISRR140 pKa = 11.84ASPNTDD146 pKa = 2.51ASALGASTLHH156 pKa = 5.83QPSPPSYY163 pKa = 8.57HH164 pKa = 6.21PHH166 pKa = 5.92YY167 pKa = 10.34AGKK170 pKa = 8.75TIDD173 pKa = 3.48FTAFAFCRR181 pKa = 11.84AVDD184 pKa = 3.71VMVITLWTRR193 pKa = 11.84TRR195 pKa = 11.84TRR197 pKa = 11.84SWHH200 pKa = 5.7PEE202 pKa = 3.32QRR204 pKa = 11.84NPRR207 pKa = 11.84LSNLVRR213 pKa = 11.84KK214 pKa = 9.02LADD217 pKa = 3.54PSVFAGSAAIIMWSWFYY234 pKa = 11.36SPEE237 pKa = 3.93RR238 pKa = 11.84LPRR241 pKa = 11.84SYY243 pKa = 11.04NQWISKK249 pKa = 10.07AADD252 pKa = 3.12IDD254 pKa = 3.64PRR256 pKa = 11.84LIQALRR262 pKa = 11.84LARR265 pKa = 11.84QGNFVYY271 pKa = 11.02GEE273 pKa = 4.2DD274 pKa = 3.47TGQAPLLAGLCRR286 pKa = 11.84EE287 pKa = 4.19LDD289 pKa = 3.8LPEE292 pKa = 4.69EE293 pKa = 4.33FADD296 pKa = 4.0PSKK299 pKa = 9.97TIPIPCEE306 pKa = 3.86LYY308 pKa = 9.99HH309 pKa = 6.54CGSGKK314 pKa = 9.94SCEE317 pKa = 3.69IHH319 pKa = 6.09ALSRR323 pKa = 11.84FWRR326 pKa = 11.84SWKK329 pKa = 9.96FAMEE333 pKa = 4.26IYY335 pKa = 10.71LPLQLLTILRR345 pKa = 11.84SPRR348 pKa = 11.84PKK350 pKa = 10.44SVLKK354 pKa = 10.54VLQAASRR361 pKa = 11.84SSSFLSGFVASFYY374 pKa = 10.97YY375 pKa = 10.16AVCLARR381 pKa = 11.84TRR383 pKa = 11.84LGPKK387 pKa = 9.43IFSQRR392 pKa = 11.84TVTPQMWDD400 pKa = 2.58SGLCVLTGCLACGWSVLLEE419 pKa = 4.3KK420 pKa = 10.34PSRR423 pKa = 11.84RR424 pKa = 11.84QEE426 pKa = 3.51IAFFVAPRR434 pKa = 11.84ALATILPRR442 pKa = 11.84VYY444 pKa = 10.06DD445 pKa = 3.07QQYY448 pKa = 9.05RR449 pKa = 11.84HH450 pKa = 6.41RR451 pKa = 11.84EE452 pKa = 3.65QAIFATSIAVVLTALKK468 pKa = 10.53SGNEE472 pKa = 3.73RR473 pKa = 11.84NIRR476 pKa = 11.84GVLGRR481 pKa = 11.84ILGGILKK488 pKa = 10.22DD489 pKa = 3.5
Molecular weight: 54.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5948075 |
39 |
6060 |
492.0 |
54.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.745 ± 0.019 | 1.224 ± 0.008 |
5.71 ± 0.015 | 6.091 ± 0.023 |
3.727 ± 0.014 | 6.879 ± 0.02 |
2.47 ± 0.011 | 4.823 ± 0.013 |
4.653 ± 0.019 | 8.972 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.17 ± 0.007 | 3.528 ± 0.01 |
6.107 ± 0.022 | 4.121 ± 0.015 |
6.195 ± 0.018 | 8.065 ± 0.023 |
6.039 ± 0.017 | 6.24 ± 0.014 |
1.485 ± 0.009 | 2.756 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
