
Sheep faeces associated smacovirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus sheas2
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
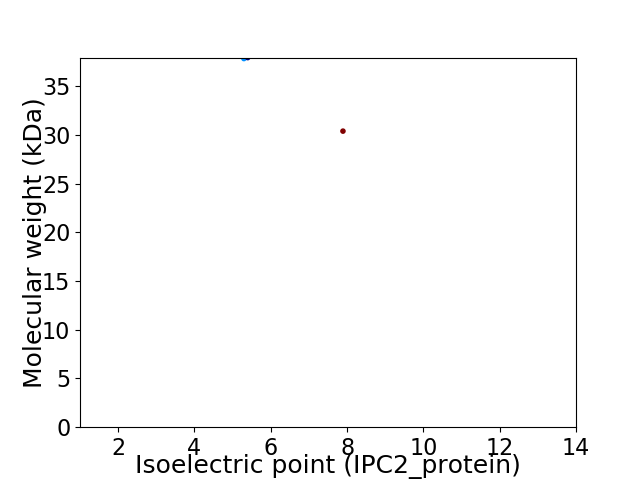
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A161J5M0|A0A161J5M0_9VIRU Putative capsid protein OS=Sheep faeces associated smacovirus 2 OX=1843757 PE=4 SV=1
MM1 pKa = 6.4TQSIAVSISEE11 pKa = 4.52TYY13 pKa = 10.73DD14 pKa = 3.7LSTTLNKK21 pKa = 9.78MGLIAIRR28 pKa = 11.84TPGMQSVEE36 pKa = 3.49KK37 pKa = 9.82RR38 pKa = 11.84YY39 pKa = 9.82PGFVRR44 pKa = 11.84NFKK47 pKa = 10.01FLRR50 pKa = 11.84VKK52 pKa = 10.59SCDD55 pKa = 3.58VVISCASSLPADD67 pKa = 4.04PLQIGTAAGSIAPQDD82 pKa = 3.58MFNPLLYY89 pKa = 10.4KK90 pKa = 10.52AVSNDD95 pKa = 2.77SWNGLLNRR103 pKa = 11.84IYY105 pKa = 10.93GSTGAVNSLGGTVRR119 pKa = 11.84FFQDD123 pKa = 2.67AFSAASSADD132 pKa = 3.76SINSYY137 pKa = 10.56YY138 pKa = 11.28ALLSDD143 pKa = 4.12PSFKK147 pKa = 10.2KK148 pKa = 10.55AHH150 pKa = 5.01VQQGLEE156 pKa = 3.94MRR158 pKa = 11.84RR159 pKa = 11.84LVPLVYY165 pKa = 10.16HH166 pKa = 6.16VLNAGGTTPNMNGATLANVDD186 pKa = 3.85EE187 pKa = 5.05LDD189 pKa = 3.68NVNAVGNNGVSVSASNNMLFGGTAFFKK216 pKa = 10.6GRR218 pKa = 11.84PVPMPPVEE226 pKa = 4.28CTTSVVHH233 pKa = 5.91PTDD236 pKa = 3.24GTVTQTVFNPIIGNIPSTYY255 pKa = 9.35VAAIVTPPATLNITYY270 pKa = 10.0FRR272 pKa = 11.84MTVRR276 pKa = 11.84WNLEE280 pKa = 3.92FFGVASDD287 pKa = 3.34IAKK290 pKa = 10.73ALMPGTSDD298 pKa = 2.64IGRR301 pKa = 11.84YY302 pKa = 9.58AYY304 pKa = 10.3FSSNVSQQTLSASKK318 pKa = 10.32ISDD321 pKa = 3.48VSEE324 pKa = 4.08TAVNADD330 pKa = 3.93DD331 pKa = 4.63QEE333 pKa = 5.72HH334 pKa = 7.22DD335 pKa = 3.22IQRR338 pKa = 11.84TVEE341 pKa = 3.99ADD343 pKa = 3.45GVTLDD348 pKa = 5.77LIMEE352 pKa = 4.61KK353 pKa = 10.72
MM1 pKa = 6.4TQSIAVSISEE11 pKa = 4.52TYY13 pKa = 10.73DD14 pKa = 3.7LSTTLNKK21 pKa = 9.78MGLIAIRR28 pKa = 11.84TPGMQSVEE36 pKa = 3.49KK37 pKa = 9.82RR38 pKa = 11.84YY39 pKa = 9.82PGFVRR44 pKa = 11.84NFKK47 pKa = 10.01FLRR50 pKa = 11.84VKK52 pKa = 10.59SCDD55 pKa = 3.58VVISCASSLPADD67 pKa = 4.04PLQIGTAAGSIAPQDD82 pKa = 3.58MFNPLLYY89 pKa = 10.4KK90 pKa = 10.52AVSNDD95 pKa = 2.77SWNGLLNRR103 pKa = 11.84IYY105 pKa = 10.93GSTGAVNSLGGTVRR119 pKa = 11.84FFQDD123 pKa = 2.67AFSAASSADD132 pKa = 3.76SINSYY137 pKa = 10.56YY138 pKa = 11.28ALLSDD143 pKa = 4.12PSFKK147 pKa = 10.2KK148 pKa = 10.55AHH150 pKa = 5.01VQQGLEE156 pKa = 3.94MRR158 pKa = 11.84RR159 pKa = 11.84LVPLVYY165 pKa = 10.16HH166 pKa = 6.16VLNAGGTTPNMNGATLANVDD186 pKa = 3.85EE187 pKa = 5.05LDD189 pKa = 3.68NVNAVGNNGVSVSASNNMLFGGTAFFKK216 pKa = 10.6GRR218 pKa = 11.84PVPMPPVEE226 pKa = 4.28CTTSVVHH233 pKa = 5.91PTDD236 pKa = 3.24GTVTQTVFNPIIGNIPSTYY255 pKa = 9.35VAAIVTPPATLNITYY270 pKa = 10.0FRR272 pKa = 11.84MTVRR276 pKa = 11.84WNLEE280 pKa = 3.92FFGVASDD287 pKa = 3.34IAKK290 pKa = 10.73ALMPGTSDD298 pKa = 2.64IGRR301 pKa = 11.84YY302 pKa = 9.58AYY304 pKa = 10.3FSSNVSQQTLSASKK318 pKa = 10.32ISDD321 pKa = 3.48VSEE324 pKa = 4.08TAVNADD330 pKa = 3.93DD331 pKa = 4.63QEE333 pKa = 5.72HH334 pKa = 7.22DD335 pKa = 3.22IQRR338 pKa = 11.84TVEE341 pKa = 3.99ADD343 pKa = 3.45GVTLDD348 pKa = 5.77LIMEE352 pKa = 4.61KK353 pKa = 10.72
Molecular weight: 37.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A161J5M0|A0A161J5M0_9VIRU Putative capsid protein OS=Sheep faeces associated smacovirus 2 OX=1843757 PE=4 SV=1
MM1 pKa = 6.36MTIPRR6 pKa = 11.84RR7 pKa = 11.84IEE9 pKa = 3.83KK10 pKa = 10.16PLDD13 pKa = 3.18NWCWQCEE20 pKa = 4.26RR21 pKa = 11.84ICSKK25 pKa = 10.99KK26 pKa = 9.26LLSYY30 pKa = 10.43IFEE33 pKa = 4.09QADD36 pKa = 3.36VKK38 pKa = 10.82RR39 pKa = 11.84YY40 pKa = 9.3IVAIEE45 pKa = 3.93KK46 pKa = 10.4GKK48 pKa = 10.6NGLDD52 pKa = 3.12HH53 pKa = 6.32FQIRR57 pKa = 11.84LSCSDD62 pKa = 3.61PDD64 pKa = 3.93FFEE67 pKa = 5.83HH68 pKa = 6.38MKK70 pKa = 10.5DD71 pKa = 2.94WCEE74 pKa = 3.53WAHH77 pKa = 5.31VEE79 pKa = 5.47KK80 pKa = 11.24AEE82 pKa = 4.15TDD84 pKa = 3.25NFDD87 pKa = 3.59YY88 pKa = 10.67EE89 pKa = 4.35RR90 pKa = 11.84KK91 pKa = 9.84EE92 pKa = 3.96GRR94 pKa = 11.84FWTSDD99 pKa = 3.19DD100 pKa = 3.4TTEE103 pKa = 4.22IRR105 pKa = 11.84ICRR108 pKa = 11.84FRR110 pKa = 11.84EE111 pKa = 3.64LGGPGRR117 pKa = 11.84QWQKK121 pKa = 11.27QIMQVLKK128 pKa = 10.05KK129 pKa = 10.27QDD131 pKa = 3.05VRR133 pKa = 11.84TIDD136 pKa = 3.48VVLDD140 pKa = 3.45PVGARR145 pKa = 11.84GKK147 pKa = 9.52SHH149 pKa = 7.2FAIALWEE156 pKa = 4.24RR157 pKa = 11.84GEE159 pKa = 3.99ALVVPRR165 pKa = 11.84YY166 pKa = 10.36SCTAEE171 pKa = 3.94KK172 pKa = 10.67LSAFVCSAYY181 pKa = 10.18RR182 pKa = 11.84GRR184 pKa = 11.84KK185 pKa = 8.88IIIIDD190 pKa = 3.42IPRR193 pKa = 11.84ANKK196 pKa = 7.67PTTALYY202 pKa = 8.45EE203 pKa = 4.29TMEE206 pKa = 4.06EE207 pKa = 4.21MKK209 pKa = 10.64DD210 pKa = 3.46GLVFDD215 pKa = 4.86PRR217 pKa = 11.84YY218 pKa = 9.88SGKK221 pKa = 8.15TRR223 pKa = 11.84NIRR226 pKa = 11.84GTKK229 pKa = 10.13VLVFTNNPLDD239 pKa = 4.06LKK241 pKa = 10.85KK242 pKa = 10.67LSHH245 pKa = 7.11DD246 pKa = 2.93RR247 pKa = 11.84WNLHH251 pKa = 6.32GISVDD256 pKa = 3.46GTLTT260 pKa = 3.23
MM1 pKa = 6.36MTIPRR6 pKa = 11.84RR7 pKa = 11.84IEE9 pKa = 3.83KK10 pKa = 10.16PLDD13 pKa = 3.18NWCWQCEE20 pKa = 4.26RR21 pKa = 11.84ICSKK25 pKa = 10.99KK26 pKa = 9.26LLSYY30 pKa = 10.43IFEE33 pKa = 4.09QADD36 pKa = 3.36VKK38 pKa = 10.82RR39 pKa = 11.84YY40 pKa = 9.3IVAIEE45 pKa = 3.93KK46 pKa = 10.4GKK48 pKa = 10.6NGLDD52 pKa = 3.12HH53 pKa = 6.32FQIRR57 pKa = 11.84LSCSDD62 pKa = 3.61PDD64 pKa = 3.93FFEE67 pKa = 5.83HH68 pKa = 6.38MKK70 pKa = 10.5DD71 pKa = 2.94WCEE74 pKa = 3.53WAHH77 pKa = 5.31VEE79 pKa = 5.47KK80 pKa = 11.24AEE82 pKa = 4.15TDD84 pKa = 3.25NFDD87 pKa = 3.59YY88 pKa = 10.67EE89 pKa = 4.35RR90 pKa = 11.84KK91 pKa = 9.84EE92 pKa = 3.96GRR94 pKa = 11.84FWTSDD99 pKa = 3.19DD100 pKa = 3.4TTEE103 pKa = 4.22IRR105 pKa = 11.84ICRR108 pKa = 11.84FRR110 pKa = 11.84EE111 pKa = 3.64LGGPGRR117 pKa = 11.84QWQKK121 pKa = 11.27QIMQVLKK128 pKa = 10.05KK129 pKa = 10.27QDD131 pKa = 3.05VRR133 pKa = 11.84TIDD136 pKa = 3.48VVLDD140 pKa = 3.45PVGARR145 pKa = 11.84GKK147 pKa = 9.52SHH149 pKa = 7.2FAIALWEE156 pKa = 4.24RR157 pKa = 11.84GEE159 pKa = 3.99ALVVPRR165 pKa = 11.84YY166 pKa = 10.36SCTAEE171 pKa = 3.94KK172 pKa = 10.67LSAFVCSAYY181 pKa = 10.18RR182 pKa = 11.84GRR184 pKa = 11.84KK185 pKa = 8.88IIIIDD190 pKa = 3.42IPRR193 pKa = 11.84ANKK196 pKa = 7.67PTTALYY202 pKa = 8.45EE203 pKa = 4.29TMEE206 pKa = 4.06EE207 pKa = 4.21MKK209 pKa = 10.64DD210 pKa = 3.46GLVFDD215 pKa = 4.86PRR217 pKa = 11.84YY218 pKa = 9.88SGKK221 pKa = 8.15TRR223 pKa = 11.84NIRR226 pKa = 11.84GTKK229 pKa = 10.13VLVFTNNPLDD239 pKa = 4.06LKK241 pKa = 10.85KK242 pKa = 10.67LSHH245 pKa = 7.11DD246 pKa = 2.93RR247 pKa = 11.84WNLHH251 pKa = 6.32GISVDD256 pKa = 3.46GTLTT260 pKa = 3.23
Molecular weight: 30.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
613 |
260 |
353 |
306.5 |
34.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.341 ± 1.425 | 1.794 ± 0.781 |
6.199 ± 0.675 | 4.568 ± 1.434 |
4.405 ± 0.106 | 6.525 ± 0.46 |
1.631 ± 0.412 | 6.199 ± 0.675 |
5.22 ± 1.739 | 7.341 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.773 ± 0.283 | 5.22 ± 1.305 |
4.731 ± 0.539 | 3.263 ± 0.113 |
5.71 ± 1.675 | 7.667 ± 1.858 |
7.015 ± 0.758 | 7.83 ± 1.255 |
1.631 ± 0.88 | 2.936 ± 0.149 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
