
Botryosphaeria dothidea virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
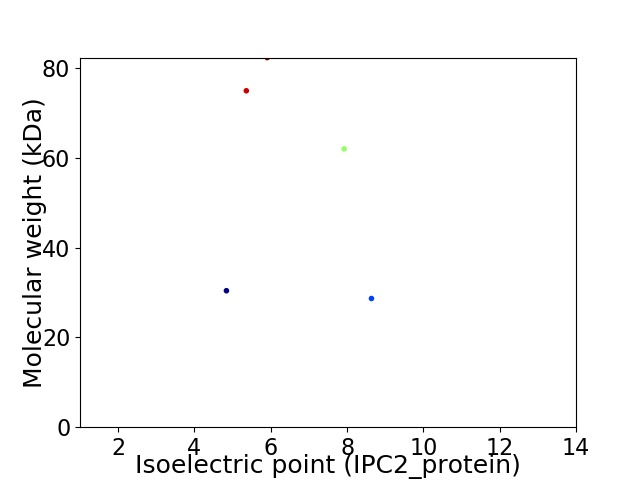
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
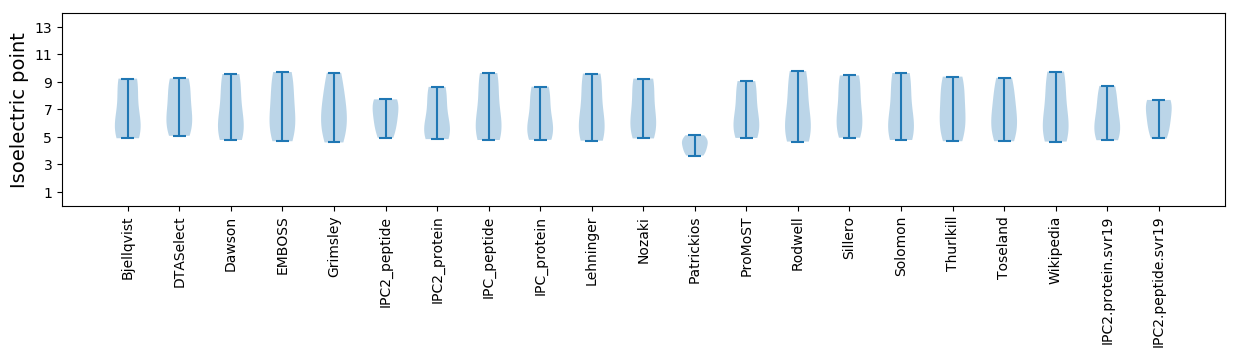
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F6TMZ8|A0A0F6TMZ8_9VIRU Capsid protein OS=Botryosphaeria dothidea virus 1 OX=1516075 GN=CP PE=4 SV=1
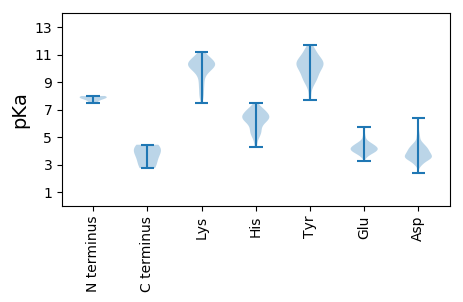
MM1 pKa = 7.86AFPMSYY7 pKa = 10.07RR8 pKa = 11.84QFIDD12 pKa = 4.24TIPDD16 pKa = 3.63EE17 pKa = 4.5IFFAYY22 pKa = 10.3AEE24 pKa = 4.46AFAQADD30 pKa = 4.07DD31 pKa = 4.3PVGLLQRR38 pKa = 11.84IRR40 pKa = 11.84EE41 pKa = 4.07ADD43 pKa = 3.5GEE45 pKa = 4.36DD46 pKa = 3.21IEE48 pKa = 5.7FEE50 pKa = 3.89ISGQLVSMPCLIDD63 pKa = 4.37CSDD66 pKa = 4.16ILDD69 pKa = 4.26GTAGTQGPSGATQEE83 pKa = 3.9IVEE86 pKa = 4.3AVEE89 pKa = 3.88EE90 pKa = 4.48HH91 pKa = 5.29VTPDD95 pKa = 2.99VGFVGTGVCAQLTIATQRR113 pKa = 11.84LYY115 pKa = 10.39PHH117 pKa = 6.65NEE119 pKa = 3.55IEE121 pKa = 4.39AVRR124 pKa = 11.84AMRR127 pKa = 11.84MDD129 pKa = 5.09DD130 pKa = 3.96GPVDD134 pKa = 3.52TSQPYY139 pKa = 9.82RR140 pKa = 11.84GVAEE144 pKa = 4.25FTGATSDD151 pKa = 3.8PHH153 pKa = 6.48PGWGGDD159 pKa = 3.9ALPAAPVFYY168 pKa = 9.74TGHH171 pKa = 6.5YY172 pKa = 9.97GPAPTCAYY180 pKa = 10.36VGDD183 pKa = 4.07GLVVFAQTHH192 pKa = 4.25THH194 pKa = 6.1LKK196 pKa = 8.41EE197 pKa = 3.99QVRR200 pKa = 11.84TAWTKK205 pKa = 10.52GPRR208 pKa = 11.84EE209 pKa = 4.08GDD211 pKa = 3.44GVSYY215 pKa = 10.49YY216 pKa = 10.88QLAKK220 pKa = 9.6TPHH223 pKa = 5.54GVKK226 pKa = 10.2AFKK229 pKa = 10.28LQYY232 pKa = 8.57VANRR236 pKa = 11.84GTLLWNGRR244 pKa = 11.84VPLGYY249 pKa = 10.42LDD251 pKa = 4.24FVKK254 pKa = 10.64QLGVIRR260 pKa = 11.84ARR262 pKa = 11.84DD263 pKa = 3.98RR264 pKa = 11.84RR265 pKa = 11.84WGPPPGSDD273 pKa = 3.76HH274 pKa = 7.09RR275 pKa = 11.84LEE277 pKa = 4.36PVV279 pKa = 2.74
MM1 pKa = 7.86AFPMSYY7 pKa = 10.07RR8 pKa = 11.84QFIDD12 pKa = 4.24TIPDD16 pKa = 3.63EE17 pKa = 4.5IFFAYY22 pKa = 10.3AEE24 pKa = 4.46AFAQADD30 pKa = 4.07DD31 pKa = 4.3PVGLLQRR38 pKa = 11.84IRR40 pKa = 11.84EE41 pKa = 4.07ADD43 pKa = 3.5GEE45 pKa = 4.36DD46 pKa = 3.21IEE48 pKa = 5.7FEE50 pKa = 3.89ISGQLVSMPCLIDD63 pKa = 4.37CSDD66 pKa = 4.16ILDD69 pKa = 4.26GTAGTQGPSGATQEE83 pKa = 3.9IVEE86 pKa = 4.3AVEE89 pKa = 3.88EE90 pKa = 4.48HH91 pKa = 5.29VTPDD95 pKa = 2.99VGFVGTGVCAQLTIATQRR113 pKa = 11.84LYY115 pKa = 10.39PHH117 pKa = 6.65NEE119 pKa = 3.55IEE121 pKa = 4.39AVRR124 pKa = 11.84AMRR127 pKa = 11.84MDD129 pKa = 5.09DD130 pKa = 3.96GPVDD134 pKa = 3.52TSQPYY139 pKa = 9.82RR140 pKa = 11.84GVAEE144 pKa = 4.25FTGATSDD151 pKa = 3.8PHH153 pKa = 6.48PGWGGDD159 pKa = 3.9ALPAAPVFYY168 pKa = 9.74TGHH171 pKa = 6.5YY172 pKa = 9.97GPAPTCAYY180 pKa = 10.36VGDD183 pKa = 4.07GLVVFAQTHH192 pKa = 4.25THH194 pKa = 6.1LKK196 pKa = 8.41EE197 pKa = 3.99QVRR200 pKa = 11.84TAWTKK205 pKa = 10.52GPRR208 pKa = 11.84EE209 pKa = 4.08GDD211 pKa = 3.44GVSYY215 pKa = 10.49YY216 pKa = 10.88QLAKK220 pKa = 9.6TPHH223 pKa = 5.54GVKK226 pKa = 10.2AFKK229 pKa = 10.28LQYY232 pKa = 8.57VANRR236 pKa = 11.84GTLLWNGRR244 pKa = 11.84VPLGYY249 pKa = 10.42LDD251 pKa = 4.24FVKK254 pKa = 10.64QLGVIRR260 pKa = 11.84ARR262 pKa = 11.84DD263 pKa = 3.98RR264 pKa = 11.84RR265 pKa = 11.84WGPPPGSDD273 pKa = 3.76HH274 pKa = 7.09RR275 pKa = 11.84LEE277 pKa = 4.36PVV279 pKa = 2.74
Molecular weight: 30.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F6TN89|A0A0F6TN89_9VIRU Uncharacterized protein OS=Botryosphaeria dothidea virus 1 OX=1516075 PE=4 SV=1
MM1 pKa = 7.99ASTTTTNNGPLLLTKK16 pKa = 10.26EE17 pKa = 4.2QASLLAALGEE27 pKa = 4.51GEE29 pKa = 4.05VAAIIKK35 pKa = 9.52LASLGLRR42 pKa = 11.84PTGILEE48 pKa = 3.84YY49 pKa = 10.88ARR51 pKa = 11.84AVDD54 pKa = 3.9GDD56 pKa = 4.43EE57 pKa = 4.79VPAPPTPNTAAKK69 pKa = 9.68PVTIVAWSFLKK80 pKa = 10.85DD81 pKa = 2.94RR82 pKa = 11.84GQYY85 pKa = 8.81HH86 pKa = 6.05ATYY89 pKa = 10.24GLSAPRR95 pKa = 11.84AGEE98 pKa = 3.97LLEE101 pKa = 5.6LLLTDD106 pKa = 4.17ADD108 pKa = 3.59AAARR112 pKa = 11.84EE113 pKa = 4.1IHH115 pKa = 6.68TIVADD120 pKa = 3.83ALRR123 pKa = 11.84QRR125 pKa = 11.84GSPRR129 pKa = 11.84PVHH132 pKa = 5.24VTLDD136 pKa = 3.76GVPSRR141 pKa = 11.84KK142 pKa = 9.44KK143 pKa = 10.63AGGEE147 pKa = 3.93GAPTGDD153 pKa = 3.77GGLGLLSQEE162 pKa = 4.22IASNPGTYY170 pKa = 10.08GSYY173 pKa = 10.37RR174 pKa = 11.84FVPEE178 pKa = 3.62QTGQPGARR186 pKa = 11.84AYY188 pKa = 10.01RR189 pKa = 11.84VRR191 pKa = 11.84LGGGLYY197 pKa = 10.54VFAQSKK203 pKa = 10.25KK204 pKa = 10.03LATAAARR211 pKa = 11.84ITRR214 pKa = 11.84VKK216 pKa = 10.5GRR218 pKa = 11.84SDD220 pKa = 3.27PLVRR224 pKa = 11.84DD225 pKa = 5.55FVAFWATGSSAKK237 pKa = 10.38FGGEE241 pKa = 3.67IPDD244 pKa = 3.41KK245 pKa = 10.4VTFDD249 pKa = 3.52GRR251 pKa = 11.84QGPNEE256 pKa = 4.22PLQSDD261 pKa = 4.12PTTKK265 pKa = 10.75AKK267 pKa = 7.46TTAGPSSEE275 pKa = 4.04
MM1 pKa = 7.99ASTTTTNNGPLLLTKK16 pKa = 10.26EE17 pKa = 4.2QASLLAALGEE27 pKa = 4.51GEE29 pKa = 4.05VAAIIKK35 pKa = 9.52LASLGLRR42 pKa = 11.84PTGILEE48 pKa = 3.84YY49 pKa = 10.88ARR51 pKa = 11.84AVDD54 pKa = 3.9GDD56 pKa = 4.43EE57 pKa = 4.79VPAPPTPNTAAKK69 pKa = 9.68PVTIVAWSFLKK80 pKa = 10.85DD81 pKa = 2.94RR82 pKa = 11.84GQYY85 pKa = 8.81HH86 pKa = 6.05ATYY89 pKa = 10.24GLSAPRR95 pKa = 11.84AGEE98 pKa = 3.97LLEE101 pKa = 5.6LLLTDD106 pKa = 4.17ADD108 pKa = 3.59AAARR112 pKa = 11.84EE113 pKa = 4.1IHH115 pKa = 6.68TIVADD120 pKa = 3.83ALRR123 pKa = 11.84QRR125 pKa = 11.84GSPRR129 pKa = 11.84PVHH132 pKa = 5.24VTLDD136 pKa = 3.76GVPSRR141 pKa = 11.84KK142 pKa = 9.44KK143 pKa = 10.63AGGEE147 pKa = 3.93GAPTGDD153 pKa = 3.77GGLGLLSQEE162 pKa = 4.22IASNPGTYY170 pKa = 10.08GSYY173 pKa = 10.37RR174 pKa = 11.84FVPEE178 pKa = 3.62QTGQPGARR186 pKa = 11.84AYY188 pKa = 10.01RR189 pKa = 11.84VRR191 pKa = 11.84LGGGLYY197 pKa = 10.54VFAQSKK203 pKa = 10.25KK204 pKa = 10.03LATAAARR211 pKa = 11.84ITRR214 pKa = 11.84VKK216 pKa = 10.5GRR218 pKa = 11.84SDD220 pKa = 3.27PLVRR224 pKa = 11.84DD225 pKa = 5.55FVAFWATGSSAKK237 pKa = 10.38FGGEE241 pKa = 3.67IPDD244 pKa = 3.41KK245 pKa = 10.4VTFDD249 pKa = 3.52GRR251 pKa = 11.84QGPNEE256 pKa = 4.22PLQSDD261 pKa = 4.12PTTKK265 pKa = 10.75AKK267 pKa = 7.46TTAGPSSEE275 pKa = 4.04
Molecular weight: 28.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2577 |
275 |
757 |
515.4 |
55.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.418 ± 0.437 | 0.97 ± 0.264 |
6.364 ± 0.329 | 5.2 ± 0.138 |
3.221 ± 0.242 | 7.916 ± 0.695 |
2.445 ± 0.188 | 3.454 ± 0.206 |
2.716 ± 0.452 | 9.313 ± 0.627 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.289 ± 0.306 | 2.212 ± 0.158 |
6.17 ± 0.476 | 2.872 ± 0.294 |
7.683 ± 0.323 | 6.558 ± 0.582 |
6.403 ± 0.467 | 8.071 ± 0.805 |
0.854 ± 0.116 | 2.833 ± 0.204 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
