
Columbid alphaherpesvirus 1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Mardivirus
Average proteome isoelectric point is 7.15
Get precalculated fractions of proteins
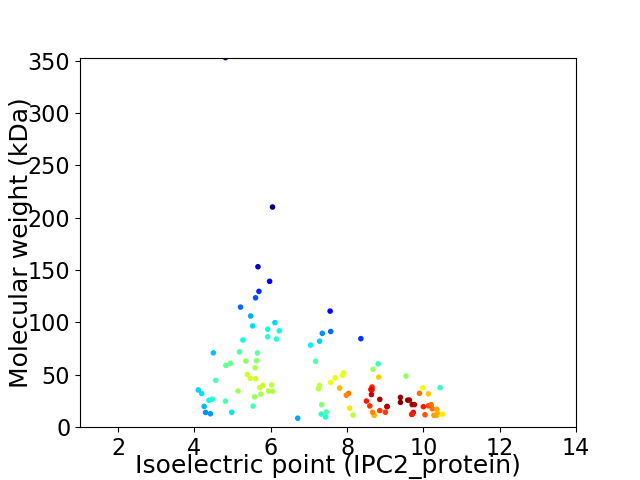
Virtual 2D-PAGE plot for 108 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V0M8E1|A0A1V0M8E1_9ALPH Uracil-DNA glycosylase OS=Columbid alphaherpesvirus 1 OX=93386 GN=CoHVHLJ_011 PE=3 SV=1
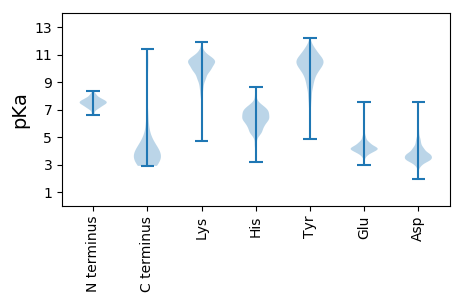
MM1 pKa = 7.49SSAIATFITYY11 pKa = 8.14TFVGAKK17 pKa = 8.9TSPTWTVPNYY27 pKa = 9.34EE28 pKa = 4.67QIICSCDD35 pKa = 2.68GGTRR39 pKa = 11.84SVAVGGMSRR48 pKa = 11.84CDD50 pKa = 3.54YY51 pKa = 11.18LPATNVILQRR61 pKa = 11.84GPAGTLLILDD71 pKa = 4.67GGVEE75 pKa = 4.34FCSYY79 pKa = 11.2LLRR82 pKa = 11.84SEE84 pKa = 4.9DD85 pKa = 3.43ATSARR90 pKa = 11.84SSGYY94 pKa = 10.26LSSAPGSICVVPFSSCTVIGLDD116 pKa = 3.77TYY118 pKa = 11.38VCNNSSGVLTVTWLRR133 pKa = 11.84QAAYY137 pKa = 8.21ITLTVYY143 pKa = 10.89GSGHH147 pKa = 6.49EE148 pKa = 4.13YY149 pKa = 10.42RR150 pKa = 11.84ISEE153 pKa = 4.56PGSVSQSSCSSVASQKK169 pKa = 11.14DD170 pKa = 3.49SAAEE174 pKa = 4.04SCEE177 pKa = 3.62GRR179 pKa = 11.84AVPTPEE185 pKa = 4.33NRR187 pKa = 11.84VPDD190 pKa = 4.24DD191 pKa = 3.59AVAEE195 pKa = 4.23ARR197 pKa = 11.84DD198 pKa = 3.84GASAPRR204 pKa = 11.84EE205 pKa = 3.97PDD207 pKa = 3.31DD208 pKa = 4.03VLDD211 pKa = 6.77EE212 pKa = 4.67IIQWNVHH219 pKa = 5.15GPGAGLAGALSPGSTPTQTKK239 pKa = 9.81NGEE242 pKa = 4.15GGGDD246 pKa = 3.56LLAAAMRR253 pKa = 11.84EE254 pKa = 4.03SHH256 pKa = 6.9LGVEE260 pKa = 4.63PLSFSDD266 pKa = 4.89IDD268 pKa = 3.52VDD270 pKa = 5.09LPNIPIPLPAQSPPAIAPVEE290 pKa = 4.3YY291 pKa = 9.98IFQDD295 pKa = 3.07NDD297 pKa = 2.87QPADD301 pKa = 3.86FGFLSSGPP309 pKa = 3.69
MM1 pKa = 7.49SSAIATFITYY11 pKa = 8.14TFVGAKK17 pKa = 8.9TSPTWTVPNYY27 pKa = 9.34EE28 pKa = 4.67QIICSCDD35 pKa = 2.68GGTRR39 pKa = 11.84SVAVGGMSRR48 pKa = 11.84CDD50 pKa = 3.54YY51 pKa = 11.18LPATNVILQRR61 pKa = 11.84GPAGTLLILDD71 pKa = 4.67GGVEE75 pKa = 4.34FCSYY79 pKa = 11.2LLRR82 pKa = 11.84SEE84 pKa = 4.9DD85 pKa = 3.43ATSARR90 pKa = 11.84SSGYY94 pKa = 10.26LSSAPGSICVVPFSSCTVIGLDD116 pKa = 3.77TYY118 pKa = 11.38VCNNSSGVLTVTWLRR133 pKa = 11.84QAAYY137 pKa = 8.21ITLTVYY143 pKa = 10.89GSGHH147 pKa = 6.49EE148 pKa = 4.13YY149 pKa = 10.42RR150 pKa = 11.84ISEE153 pKa = 4.56PGSVSQSSCSSVASQKK169 pKa = 11.14DD170 pKa = 3.49SAAEE174 pKa = 4.04SCEE177 pKa = 3.62GRR179 pKa = 11.84AVPTPEE185 pKa = 4.33NRR187 pKa = 11.84VPDD190 pKa = 4.24DD191 pKa = 3.59AVAEE195 pKa = 4.23ARR197 pKa = 11.84DD198 pKa = 3.84GASAPRR204 pKa = 11.84EE205 pKa = 3.97PDD207 pKa = 3.31DD208 pKa = 4.03VLDD211 pKa = 6.77EE212 pKa = 4.67IIQWNVHH219 pKa = 5.15GPGAGLAGALSPGSTPTQTKK239 pKa = 9.81NGEE242 pKa = 4.15GGGDD246 pKa = 3.56LLAAAMRR253 pKa = 11.84EE254 pKa = 4.03SHH256 pKa = 6.9LGVEE260 pKa = 4.63PLSFSDD266 pKa = 4.89IDD268 pKa = 3.52VDD270 pKa = 5.09LPNIPIPLPAQSPPAIAPVEE290 pKa = 4.3YY291 pKa = 9.98IFQDD295 pKa = 3.07NDD297 pKa = 2.87QPADD301 pKa = 3.86FGFLSSGPP309 pKa = 3.69
Molecular weight: 32.11 kDa
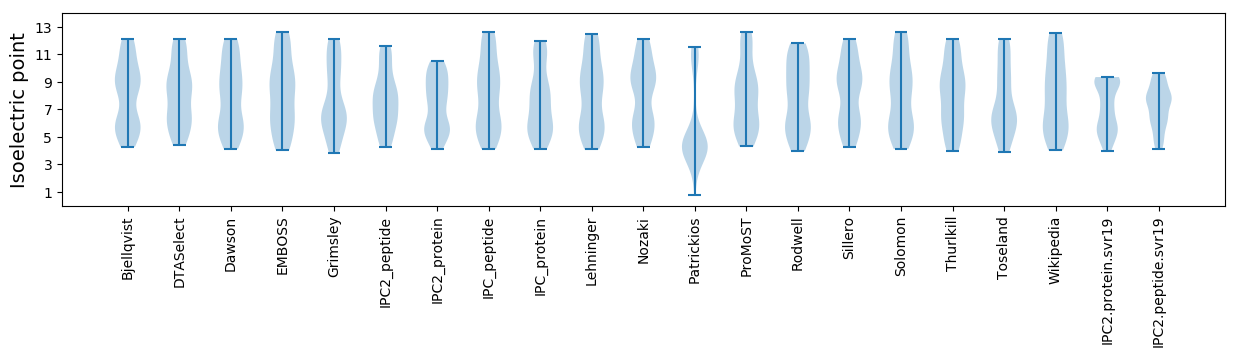
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V0M8M6|A0A1V0M8M6_9ALPH Virion protein US2 OS=Columbid alphaherpesvirus 1 OX=93386 GN=CoHVHLJ_106 PE=4 SV=1
MM1 pKa = 7.64GLPLRR6 pKa = 11.84RR7 pKa = 11.84SSRR10 pKa = 11.84IRR12 pKa = 11.84GAAGHH17 pKa = 6.12VATALVSRR25 pKa = 11.84LEE27 pKa = 3.92TAARR31 pKa = 11.84YY32 pKa = 9.48ASGGRR37 pKa = 11.84RR38 pKa = 11.84PVTRR42 pKa = 11.84SCAVGRR48 pKa = 11.84LHH50 pKa = 7.31LGVGINNRR58 pKa = 11.84RR59 pKa = 11.84VRR61 pKa = 11.84GGPQTSPSAASQPASAIPASRR82 pKa = 11.84RR83 pKa = 11.84TALGMRR89 pKa = 11.84GVGGVIGEE97 pKa = 4.19VAKK100 pKa = 10.54RR101 pKa = 11.84VAGGEE106 pKa = 4.12RR107 pKa = 11.84AASAPRR113 pKa = 11.84ARR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84PRR119 pKa = 11.84SSLNSNAAAAAAAAAATGSDD139 pKa = 4.05APQMVPGQTKK149 pKa = 9.94KK150 pKa = 10.73CGAPKK155 pKa = 10.35RR156 pKa = 11.84STAATTTTTATTADD170 pKa = 4.38GGCSCAGGGAACGAHH185 pKa = 5.78HH186 pKa = 7.35RR187 pKa = 11.84EE188 pKa = 3.98RR189 pKa = 11.84DD190 pKa = 3.12GHH192 pKa = 5.11RR193 pKa = 11.84RR194 pKa = 11.84RR195 pKa = 11.84TTVCMGTCAIGRR207 pKa = 11.84RR208 pKa = 11.84RR209 pKa = 11.84SDD211 pKa = 2.8ASRR214 pKa = 11.84EE215 pKa = 4.31CACMPPPASTTPAGAGAVAAAAVAPCVDD243 pKa = 3.46SRR245 pKa = 11.84TLRR248 pKa = 11.84PQQRR252 pKa = 11.84PHH254 pKa = 5.73PRR256 pKa = 11.84SRR258 pKa = 11.84RR259 pKa = 11.84GTVSPIEE266 pKa = 4.16ASVASPAGPSDD277 pKa = 3.29ADD279 pKa = 3.17YY280 pKa = 11.32RR281 pKa = 11.84RR282 pKa = 11.84LYY284 pKa = 10.29KK285 pKa = 10.52CRR287 pKa = 11.84LDD289 pKa = 4.07TEE291 pKa = 4.18EE292 pKa = 4.87HH293 pKa = 6.18AASLPRR299 pKa = 11.84SVKK302 pKa = 10.16DD303 pKa = 3.34LAHH306 pKa = 6.26ITHH309 pKa = 6.81TAARR313 pKa = 11.84DD314 pKa = 3.33AYY316 pKa = 9.04SAMRR320 pKa = 11.84AGMPPPARR328 pKa = 11.84LWKK331 pKa = 10.32IVGCKK336 pKa = 9.98FKK338 pKa = 11.12ALLQSYY344 pKa = 8.19EE345 pKa = 4.0RR346 pKa = 11.84SAPPFNPSEE355 pKa = 4.63PIRR358 pKa = 11.84TVAAKK363 pKa = 10.35AFRR366 pKa = 11.84AIAAVPPASHH376 pKa = 6.5QEE378 pKa = 3.86LSDD381 pKa = 3.52ALTLVMYY388 pKa = 7.4WCCLGHH394 pKa = 6.77VSRR397 pKa = 11.84CKK399 pKa = 9.78MAGLYY404 pKa = 9.38EE405 pKa = 4.32RR406 pKa = 11.84EE407 pKa = 4.09PAVPALFDD415 pKa = 3.73ARR417 pKa = 11.84PGCGEE422 pKa = 4.26IPPSDD427 pKa = 3.51VNYY430 pKa = 9.64WKK432 pKa = 10.26QLYY435 pKa = 9.96RR436 pKa = 11.84LADD439 pKa = 3.43KK440 pKa = 10.65GGSSIVPDD448 pKa = 4.06ASLAAYY454 pKa = 8.75VALVRR459 pKa = 11.84RR460 pKa = 11.84DD461 pKa = 3.49PDD463 pKa = 3.47EE464 pKa = 4.26EE465 pKa = 4.28LVGG468 pKa = 3.88
MM1 pKa = 7.64GLPLRR6 pKa = 11.84RR7 pKa = 11.84SSRR10 pKa = 11.84IRR12 pKa = 11.84GAAGHH17 pKa = 6.12VATALVSRR25 pKa = 11.84LEE27 pKa = 3.92TAARR31 pKa = 11.84YY32 pKa = 9.48ASGGRR37 pKa = 11.84RR38 pKa = 11.84PVTRR42 pKa = 11.84SCAVGRR48 pKa = 11.84LHH50 pKa = 7.31LGVGINNRR58 pKa = 11.84RR59 pKa = 11.84VRR61 pKa = 11.84GGPQTSPSAASQPASAIPASRR82 pKa = 11.84RR83 pKa = 11.84TALGMRR89 pKa = 11.84GVGGVIGEE97 pKa = 4.19VAKK100 pKa = 10.54RR101 pKa = 11.84VAGGEE106 pKa = 4.12RR107 pKa = 11.84AASAPRR113 pKa = 11.84ARR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84PRR119 pKa = 11.84SSLNSNAAAAAAAAAATGSDD139 pKa = 4.05APQMVPGQTKK149 pKa = 9.94KK150 pKa = 10.73CGAPKK155 pKa = 10.35RR156 pKa = 11.84STAATTTTTATTADD170 pKa = 4.38GGCSCAGGGAACGAHH185 pKa = 5.78HH186 pKa = 7.35RR187 pKa = 11.84EE188 pKa = 3.98RR189 pKa = 11.84DD190 pKa = 3.12GHH192 pKa = 5.11RR193 pKa = 11.84RR194 pKa = 11.84RR195 pKa = 11.84TTVCMGTCAIGRR207 pKa = 11.84RR208 pKa = 11.84RR209 pKa = 11.84SDD211 pKa = 2.8ASRR214 pKa = 11.84EE215 pKa = 4.31CACMPPPASTTPAGAGAVAAAAVAPCVDD243 pKa = 3.46SRR245 pKa = 11.84TLRR248 pKa = 11.84PQQRR252 pKa = 11.84PHH254 pKa = 5.73PRR256 pKa = 11.84SRR258 pKa = 11.84RR259 pKa = 11.84GTVSPIEE266 pKa = 4.16ASVASPAGPSDD277 pKa = 3.29ADD279 pKa = 3.17YY280 pKa = 11.32RR281 pKa = 11.84RR282 pKa = 11.84LYY284 pKa = 10.29KK285 pKa = 10.52CRR287 pKa = 11.84LDD289 pKa = 4.07TEE291 pKa = 4.18EE292 pKa = 4.87HH293 pKa = 6.18AASLPRR299 pKa = 11.84SVKK302 pKa = 10.16DD303 pKa = 3.34LAHH306 pKa = 6.26ITHH309 pKa = 6.81TAARR313 pKa = 11.84DD314 pKa = 3.33AYY316 pKa = 9.04SAMRR320 pKa = 11.84AGMPPPARR328 pKa = 11.84LWKK331 pKa = 10.32IVGCKK336 pKa = 9.98FKK338 pKa = 11.12ALLQSYY344 pKa = 8.19EE345 pKa = 4.0RR346 pKa = 11.84SAPPFNPSEE355 pKa = 4.63PIRR358 pKa = 11.84TVAAKK363 pKa = 10.35AFRR366 pKa = 11.84AIAAVPPASHH376 pKa = 6.5QEE378 pKa = 3.86LSDD381 pKa = 3.52ALTLVMYY388 pKa = 7.4WCCLGHH394 pKa = 6.77VSRR397 pKa = 11.84CKK399 pKa = 9.78MAGLYY404 pKa = 9.38EE405 pKa = 4.32RR406 pKa = 11.84EE407 pKa = 4.09PAVPALFDD415 pKa = 3.73ARR417 pKa = 11.84PGCGEE422 pKa = 4.26IPPSDD427 pKa = 3.51VNYY430 pKa = 9.64WKK432 pKa = 10.26QLYY435 pKa = 9.96RR436 pKa = 11.84LADD439 pKa = 3.43KK440 pKa = 10.65GGSSIVPDD448 pKa = 4.06ASLAAYY454 pKa = 8.75VALVRR459 pKa = 11.84RR460 pKa = 11.84DD461 pKa = 3.49PDD463 pKa = 3.47EE464 pKa = 4.26EE465 pKa = 4.28LVGG468 pKa = 3.88
Molecular weight: 48.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
47845 |
77 |
3280 |
443.0 |
48.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.066 ± 0.359 | 2.067 ± 0.154 |
6.239 ± 0.201 | 5.449 ± 0.194 |
3.424 ± 0.156 | 7.719 ± 0.252 |
1.91 ± 0.08 | 3.944 ± 0.132 |
3.334 ± 0.158 | 8.523 ± 0.202 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.278 ± 0.081 | 2.799 ± 0.134 |
6.191 ± 0.248 | 2.558 ± 0.098 |
8.663 ± 0.258 | 8.206 ± 0.241 |
5.792 ± 0.14 | 6.749 ± 0.157 |
1.158 ± 0.074 | 2.932 ± 0.116 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
