
Rhizoctonia solani dsRNA virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
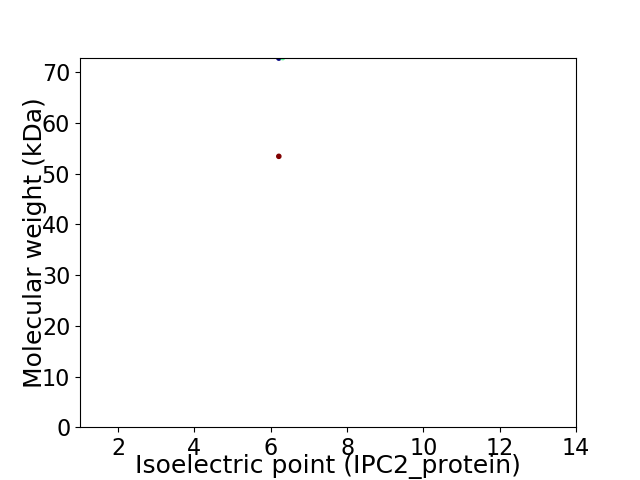
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U5QCB6|U5QCB6_9VIRU Coat protein OS=Rhizoctonia solani dsRNA virus 2 OX=1411681 PE=4 SV=1
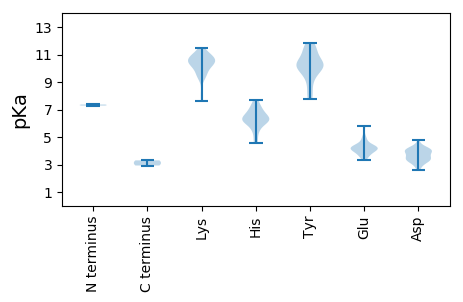
MM1 pKa = 7.29NLYY4 pKa = 10.38NRR6 pKa = 11.84VSALFANWFSSPSNLEE22 pKa = 3.87FVGSYY27 pKa = 9.04HH28 pKa = 6.16HH29 pKa = 6.77QPGTVPPNPSTQEE42 pKa = 3.36AHH44 pKa = 7.15KK45 pKa = 10.6RR46 pKa = 11.84FLHH49 pKa = 6.15NVFKK53 pKa = 10.89QHH55 pKa = 6.36LFTYY59 pKa = 9.56EE60 pKa = 3.47LDD62 pKa = 4.05YY63 pKa = 11.34IEE65 pKa = 4.94NEE67 pKa = 3.91HH68 pKa = 6.46RR69 pKa = 11.84RR70 pKa = 11.84SEE72 pKa = 4.15ATPEE76 pKa = 4.39AIEE79 pKa = 4.26NDD81 pKa = 3.52FFANDD86 pKa = 3.98VEE88 pKa = 4.33PHH90 pKa = 7.16DD91 pKa = 4.15IPFDD95 pKa = 3.5VHH97 pKa = 7.54VEE99 pKa = 3.69IGLQCMTDD107 pKa = 3.13AFRR110 pKa = 11.84PPVPCLPAHH119 pKa = 6.22LNDD122 pKa = 4.24VEE124 pKa = 4.12HH125 pKa = 7.16HH126 pKa = 6.92YY127 pKa = 10.22PFKK130 pKa = 9.92WQVNAEE136 pKa = 4.34PPFSTDD142 pKa = 4.82SYY144 pKa = 11.46FLDD147 pKa = 3.46NRR149 pKa = 11.84KK150 pKa = 9.51LFSDD154 pKa = 4.4YY155 pKa = 11.24YY156 pKa = 10.09DD157 pKa = 3.89TEE159 pKa = 4.26SQTWRR164 pKa = 11.84GYY166 pKa = 10.53VDD168 pKa = 3.3PFEE171 pKa = 4.88ANRR174 pKa = 11.84RR175 pKa = 11.84YY176 pKa = 9.77QHH178 pKa = 6.17TKK180 pKa = 10.52DD181 pKa = 3.82KK182 pKa = 11.29EE183 pKa = 4.21GFLNQTVPAKK193 pKa = 10.36FGFMKK198 pKa = 9.96DD199 pKa = 3.91TIFSWTRR206 pKa = 11.84RR207 pKa = 11.84WHH209 pKa = 6.41HH210 pKa = 6.45IIKK213 pKa = 10.42EE214 pKa = 4.27GFQTATNLSSTAYY227 pKa = 10.47LRR229 pKa = 11.84DD230 pKa = 3.33RR231 pKa = 11.84FIFPMLLHH239 pKa = 6.66TKK241 pKa = 8.25TAIVKK246 pKa = 10.27KK247 pKa = 10.36DD248 pKa = 4.02DD249 pKa = 4.05PDD251 pKa = 3.54KK252 pKa = 11.15MRR254 pKa = 11.84TIWGCSKK261 pKa = 10.15PWIIADD267 pKa = 3.44TMFYY271 pKa = 10.05WEE273 pKa = 4.23YY274 pKa = 10.03IAWIKK279 pKa = 9.49KK280 pKa = 9.4HH281 pKa = 6.16PGVTPMLWGYY291 pKa = 7.8EE292 pKa = 4.05TMTGGWMRR300 pKa = 11.84LNSQLFSSYY309 pKa = 10.46LKK311 pKa = 10.44KK312 pKa = 10.88SYY314 pKa = 8.94VTLDD318 pKa = 2.91WSRR321 pKa = 11.84FDD323 pKa = 2.98KK324 pKa = 10.75RR325 pKa = 11.84AYY327 pKa = 10.09FKK329 pKa = 11.06LILAIMCRR337 pKa = 11.84VRR339 pKa = 11.84TFLDD343 pKa = 3.38FDD345 pKa = 3.59NGYY348 pKa = 9.79LPNVNYY354 pKa = 10.02PDD356 pKa = 4.0TRR358 pKa = 11.84TDD360 pKa = 2.92WSPNKK365 pKa = 9.93AQRR368 pKa = 11.84LEE370 pKa = 4.36RR371 pKa = 11.84LWLWTLEE378 pKa = 4.21CLIKK382 pKa = 10.85SPIVLPDD389 pKa = 2.63GRR391 pKa = 11.84MYY393 pKa = 10.25IRR395 pKa = 11.84HH396 pKa = 5.57YY397 pKa = 11.2AGIPSGLFITQLLDD411 pKa = 2.7SWYY414 pKa = 9.89NYY416 pKa = 9.38TMICTILSSIGLNPKK431 pKa = 9.44HH432 pKa = 6.79CIIKK436 pKa = 10.03VQGDD440 pKa = 3.66DD441 pKa = 3.69SIVRR445 pKa = 11.84LGVLIPPEE453 pKa = 3.67AHH455 pKa = 6.29EE456 pKa = 4.75AFLLALQSKK465 pKa = 10.73ADD467 pKa = 4.16FYY469 pKa = 11.32FKK471 pKa = 10.83AIISVDD477 pKa = 3.18KK478 pKa = 11.47SEE480 pKa = 5.8LGNSLNNRR488 pKa = 11.84EE489 pKa = 4.05VLSYY493 pKa = 10.99RR494 pKa = 11.84NYY496 pKa = 10.48NGLPRR501 pKa = 11.84RR502 pKa = 11.84DD503 pKa = 4.13EE504 pKa = 4.27IKK506 pKa = 10.2MLAQFYY512 pKa = 7.91HH513 pKa = 5.93TKK515 pKa = 10.44ARR517 pKa = 11.84NPTPEE522 pKa = 3.62IAMAQAVGFAYY533 pKa = 10.21ASCGTHH539 pKa = 6.09QRR541 pKa = 11.84VLDD544 pKa = 4.21ALEE547 pKa = 4.42HH548 pKa = 6.38VYY550 pKa = 10.52TDD552 pKa = 4.0YY553 pKa = 11.66KK554 pKa = 10.96DD555 pKa = 4.21AGYY558 pKa = 8.48TPNRR562 pKa = 11.84AGLSLVFGNSPDD574 pKa = 3.27IQLPHH579 pKa = 7.67YY580 pKa = 10.36DD581 pKa = 3.3IDD583 pKa = 3.91HH584 pKa = 6.61FPSIEE589 pKa = 4.03EE590 pKa = 3.62IKK592 pKa = 10.86RR593 pKa = 11.84FLTCNSYY600 pKa = 11.59DD601 pKa = 4.04NSVQMAKK608 pKa = 9.48SWPTSYY614 pKa = 10.68FISEE618 pKa = 3.97PCEE621 pKa = 3.91RR622 pKa = 11.84MM623 pKa = 3.3
MM1 pKa = 7.29NLYY4 pKa = 10.38NRR6 pKa = 11.84VSALFANWFSSPSNLEE22 pKa = 3.87FVGSYY27 pKa = 9.04HH28 pKa = 6.16HH29 pKa = 6.77QPGTVPPNPSTQEE42 pKa = 3.36AHH44 pKa = 7.15KK45 pKa = 10.6RR46 pKa = 11.84FLHH49 pKa = 6.15NVFKK53 pKa = 10.89QHH55 pKa = 6.36LFTYY59 pKa = 9.56EE60 pKa = 3.47LDD62 pKa = 4.05YY63 pKa = 11.34IEE65 pKa = 4.94NEE67 pKa = 3.91HH68 pKa = 6.46RR69 pKa = 11.84RR70 pKa = 11.84SEE72 pKa = 4.15ATPEE76 pKa = 4.39AIEE79 pKa = 4.26NDD81 pKa = 3.52FFANDD86 pKa = 3.98VEE88 pKa = 4.33PHH90 pKa = 7.16DD91 pKa = 4.15IPFDD95 pKa = 3.5VHH97 pKa = 7.54VEE99 pKa = 3.69IGLQCMTDD107 pKa = 3.13AFRR110 pKa = 11.84PPVPCLPAHH119 pKa = 6.22LNDD122 pKa = 4.24VEE124 pKa = 4.12HH125 pKa = 7.16HH126 pKa = 6.92YY127 pKa = 10.22PFKK130 pKa = 9.92WQVNAEE136 pKa = 4.34PPFSTDD142 pKa = 4.82SYY144 pKa = 11.46FLDD147 pKa = 3.46NRR149 pKa = 11.84KK150 pKa = 9.51LFSDD154 pKa = 4.4YY155 pKa = 11.24YY156 pKa = 10.09DD157 pKa = 3.89TEE159 pKa = 4.26SQTWRR164 pKa = 11.84GYY166 pKa = 10.53VDD168 pKa = 3.3PFEE171 pKa = 4.88ANRR174 pKa = 11.84RR175 pKa = 11.84YY176 pKa = 9.77QHH178 pKa = 6.17TKK180 pKa = 10.52DD181 pKa = 3.82KK182 pKa = 11.29EE183 pKa = 4.21GFLNQTVPAKK193 pKa = 10.36FGFMKK198 pKa = 9.96DD199 pKa = 3.91TIFSWTRR206 pKa = 11.84RR207 pKa = 11.84WHH209 pKa = 6.41HH210 pKa = 6.45IIKK213 pKa = 10.42EE214 pKa = 4.27GFQTATNLSSTAYY227 pKa = 10.47LRR229 pKa = 11.84DD230 pKa = 3.33RR231 pKa = 11.84FIFPMLLHH239 pKa = 6.66TKK241 pKa = 8.25TAIVKK246 pKa = 10.27KK247 pKa = 10.36DD248 pKa = 4.02DD249 pKa = 4.05PDD251 pKa = 3.54KK252 pKa = 11.15MRR254 pKa = 11.84TIWGCSKK261 pKa = 10.15PWIIADD267 pKa = 3.44TMFYY271 pKa = 10.05WEE273 pKa = 4.23YY274 pKa = 10.03IAWIKK279 pKa = 9.49KK280 pKa = 9.4HH281 pKa = 6.16PGVTPMLWGYY291 pKa = 7.8EE292 pKa = 4.05TMTGGWMRR300 pKa = 11.84LNSQLFSSYY309 pKa = 10.46LKK311 pKa = 10.44KK312 pKa = 10.88SYY314 pKa = 8.94VTLDD318 pKa = 2.91WSRR321 pKa = 11.84FDD323 pKa = 2.98KK324 pKa = 10.75RR325 pKa = 11.84AYY327 pKa = 10.09FKK329 pKa = 11.06LILAIMCRR337 pKa = 11.84VRR339 pKa = 11.84TFLDD343 pKa = 3.38FDD345 pKa = 3.59NGYY348 pKa = 9.79LPNVNYY354 pKa = 10.02PDD356 pKa = 4.0TRR358 pKa = 11.84TDD360 pKa = 2.92WSPNKK365 pKa = 9.93AQRR368 pKa = 11.84LEE370 pKa = 4.36RR371 pKa = 11.84LWLWTLEE378 pKa = 4.21CLIKK382 pKa = 10.85SPIVLPDD389 pKa = 2.63GRR391 pKa = 11.84MYY393 pKa = 10.25IRR395 pKa = 11.84HH396 pKa = 5.57YY397 pKa = 11.2AGIPSGLFITQLLDD411 pKa = 2.7SWYY414 pKa = 9.89NYY416 pKa = 9.38TMICTILSSIGLNPKK431 pKa = 9.44HH432 pKa = 6.79CIIKK436 pKa = 10.03VQGDD440 pKa = 3.66DD441 pKa = 3.69SIVRR445 pKa = 11.84LGVLIPPEE453 pKa = 3.67AHH455 pKa = 6.29EE456 pKa = 4.75AFLLALQSKK465 pKa = 10.73ADD467 pKa = 4.16FYY469 pKa = 11.32FKK471 pKa = 10.83AIISVDD477 pKa = 3.18KK478 pKa = 11.47SEE480 pKa = 5.8LGNSLNNRR488 pKa = 11.84EE489 pKa = 4.05VLSYY493 pKa = 10.99RR494 pKa = 11.84NYY496 pKa = 10.48NGLPRR501 pKa = 11.84RR502 pKa = 11.84DD503 pKa = 4.13EE504 pKa = 4.27IKK506 pKa = 10.2MLAQFYY512 pKa = 7.91HH513 pKa = 5.93TKK515 pKa = 10.44ARR517 pKa = 11.84NPTPEE522 pKa = 3.62IAMAQAVGFAYY533 pKa = 10.21ASCGTHH539 pKa = 6.09QRR541 pKa = 11.84VLDD544 pKa = 4.21ALEE547 pKa = 4.42HH548 pKa = 6.38VYY550 pKa = 10.52TDD552 pKa = 4.0YY553 pKa = 11.66KK554 pKa = 10.96DD555 pKa = 4.21AGYY558 pKa = 8.48TPNRR562 pKa = 11.84AGLSLVFGNSPDD574 pKa = 3.27IQLPHH579 pKa = 7.67YY580 pKa = 10.36DD581 pKa = 3.3IDD583 pKa = 3.91HH584 pKa = 6.61FPSIEE589 pKa = 4.03EE590 pKa = 3.62IKK592 pKa = 10.86RR593 pKa = 11.84FLTCNSYY600 pKa = 11.59DD601 pKa = 4.04NSVQMAKK608 pKa = 9.48SWPTSYY614 pKa = 10.68FISEE618 pKa = 3.97PCEE621 pKa = 3.91RR622 pKa = 11.84MM623 pKa = 3.3
Molecular weight: 72.82 kDa
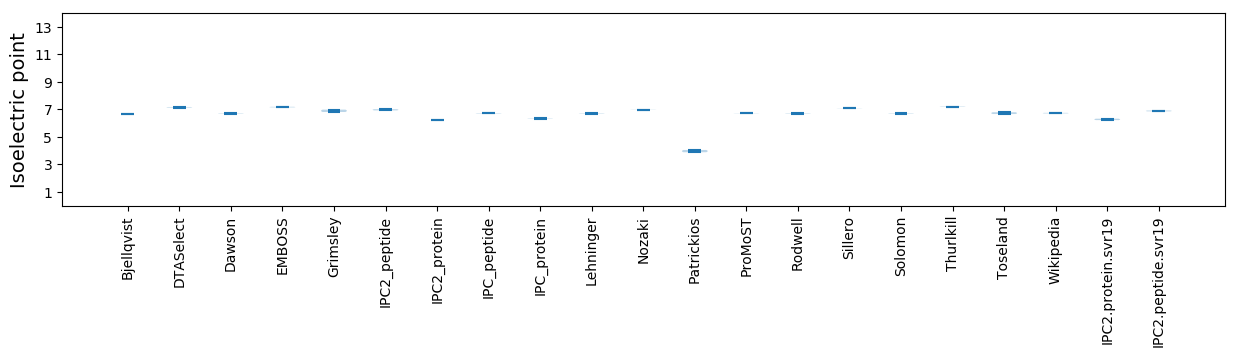
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U5QCB6|U5QCB6_9VIRU Coat protein OS=Rhizoctonia solani dsRNA virus 2 OX=1411681 PE=4 SV=1
MM1 pKa = 7.37SAKK4 pKa = 9.18TDD6 pKa = 3.67QVSVSAPDD14 pKa = 3.58AAPGNPQTPTAAAVPTAPSANPEE37 pKa = 4.14PASKK41 pKa = 10.73SSTKK45 pKa = 10.63SKK47 pKa = 10.59GVRR50 pKa = 11.84KK51 pKa = 7.63TLAFSSSSPGIAPLLNSIANVNVSIPQRR79 pKa = 11.84NQTNFFFPDD88 pKa = 3.43ATCFMRR94 pKa = 11.84CVSTCDD100 pKa = 3.28QAMSTTKK107 pKa = 10.51KK108 pKa = 10.45FLDD111 pKa = 4.12GTEE114 pKa = 3.59SWTAFVSQYY123 pKa = 10.23YY124 pKa = 10.82ASLLWHH130 pKa = 6.01LQVFRR135 pKa = 11.84TYY137 pKa = 10.59AKK139 pKa = 10.4SGIPDD144 pKa = 3.28QEE146 pKa = 4.12GQEE149 pKa = 3.94FLDD152 pKa = 4.15FFEE155 pKa = 5.72KK156 pKa = 10.61RR157 pKa = 11.84IDD159 pKa = 3.79LLNIPVPGPWIPFLRR174 pKa = 11.84SPSVVEE180 pKa = 4.05AHH182 pKa = 6.49HH183 pKa = 6.27EE184 pKa = 4.38SYY186 pKa = 11.87GNIAPVLPAIATTPLNSANFFSWPPVVSRR215 pKa = 11.84SIPNPLLPLDD225 pKa = 4.03QLHH228 pKa = 6.78ALAGHH233 pKa = 7.56AMGQNNANAFVAYY246 pKa = 8.28TNIMGMQLIGANDD259 pKa = 3.79FNNVASLLLNPTSHH273 pKa = 6.17SWSSASRR280 pKa = 11.84NRR282 pKa = 11.84TATVAEE288 pKa = 5.09FWHH291 pKa = 6.89DD292 pKa = 3.82HH293 pKa = 6.0ANMLPTRR300 pKa = 11.84LVHH303 pKa = 6.27NQAANQEE310 pKa = 4.03INTYY314 pKa = 9.26FRR316 pKa = 11.84YY317 pKa = 10.07FGFEE321 pKa = 4.3SVDD324 pKa = 3.29GTHH327 pKa = 6.25PHH329 pKa = 6.32GMLEE333 pKa = 4.27TVLNDD338 pKa = 3.1MALYY342 pKa = 10.09CQFFKK347 pKa = 11.17GSAPFSTVDD356 pKa = 3.21IVGLGASIPRR366 pKa = 11.84HH367 pKa = 5.42LPISTTAVRR376 pKa = 11.84AFVYY380 pKa = 7.9PTINRR385 pKa = 11.84LRR387 pKa = 11.84ANRR390 pKa = 11.84HH391 pKa = 4.56AQGHH395 pKa = 4.67RR396 pKa = 11.84HH397 pKa = 5.49FPSDD401 pKa = 3.27LLYY404 pKa = 11.39NMSHH408 pKa = 7.14GDD410 pKa = 3.54LTLDD414 pKa = 3.65TVGEE418 pKa = 4.22QYY420 pKa = 11.86AMSTLVNSTFGSILTQNGHH439 pKa = 5.76DD440 pKa = 4.5QIPLDD445 pKa = 4.34DD446 pKa = 3.95QVEE449 pKa = 4.69GPFWAITTMRR459 pKa = 11.84HH460 pKa = 5.22AKK462 pKa = 9.7GFSPIPSFSTVIPAYY477 pKa = 9.63YY478 pKa = 10.18HH479 pKa = 4.73VTTPVTPKK487 pKa = 10.25TSS489 pKa = 2.93
MM1 pKa = 7.37SAKK4 pKa = 9.18TDD6 pKa = 3.67QVSVSAPDD14 pKa = 3.58AAPGNPQTPTAAAVPTAPSANPEE37 pKa = 4.14PASKK41 pKa = 10.73SSTKK45 pKa = 10.63SKK47 pKa = 10.59GVRR50 pKa = 11.84KK51 pKa = 7.63TLAFSSSSPGIAPLLNSIANVNVSIPQRR79 pKa = 11.84NQTNFFFPDD88 pKa = 3.43ATCFMRR94 pKa = 11.84CVSTCDD100 pKa = 3.28QAMSTTKK107 pKa = 10.51KK108 pKa = 10.45FLDD111 pKa = 4.12GTEE114 pKa = 3.59SWTAFVSQYY123 pKa = 10.23YY124 pKa = 10.82ASLLWHH130 pKa = 6.01LQVFRR135 pKa = 11.84TYY137 pKa = 10.59AKK139 pKa = 10.4SGIPDD144 pKa = 3.28QEE146 pKa = 4.12GQEE149 pKa = 3.94FLDD152 pKa = 4.15FFEE155 pKa = 5.72KK156 pKa = 10.61RR157 pKa = 11.84IDD159 pKa = 3.79LLNIPVPGPWIPFLRR174 pKa = 11.84SPSVVEE180 pKa = 4.05AHH182 pKa = 6.49HH183 pKa = 6.27EE184 pKa = 4.38SYY186 pKa = 11.87GNIAPVLPAIATTPLNSANFFSWPPVVSRR215 pKa = 11.84SIPNPLLPLDD225 pKa = 4.03QLHH228 pKa = 6.78ALAGHH233 pKa = 7.56AMGQNNANAFVAYY246 pKa = 8.28TNIMGMQLIGANDD259 pKa = 3.79FNNVASLLLNPTSHH273 pKa = 6.17SWSSASRR280 pKa = 11.84NRR282 pKa = 11.84TATVAEE288 pKa = 5.09FWHH291 pKa = 6.89DD292 pKa = 3.82HH293 pKa = 6.0ANMLPTRR300 pKa = 11.84LVHH303 pKa = 6.27NQAANQEE310 pKa = 4.03INTYY314 pKa = 9.26FRR316 pKa = 11.84YY317 pKa = 10.07FGFEE321 pKa = 4.3SVDD324 pKa = 3.29GTHH327 pKa = 6.25PHH329 pKa = 6.32GMLEE333 pKa = 4.27TVLNDD338 pKa = 3.1MALYY342 pKa = 10.09CQFFKK347 pKa = 11.17GSAPFSTVDD356 pKa = 3.21IVGLGASIPRR366 pKa = 11.84HH367 pKa = 5.42LPISTTAVRR376 pKa = 11.84AFVYY380 pKa = 7.9PTINRR385 pKa = 11.84LRR387 pKa = 11.84ANRR390 pKa = 11.84HH391 pKa = 4.56AQGHH395 pKa = 4.67RR396 pKa = 11.84HH397 pKa = 5.49FPSDD401 pKa = 3.27LLYY404 pKa = 11.39NMSHH408 pKa = 7.14GDD410 pKa = 3.54LTLDD414 pKa = 3.65TVGEE418 pKa = 4.22QYY420 pKa = 11.86AMSTLVNSTFGSILTQNGHH439 pKa = 5.76DD440 pKa = 4.5QIPLDD445 pKa = 4.34DD446 pKa = 3.95QVEE449 pKa = 4.69GPFWAITTMRR459 pKa = 11.84HH460 pKa = 5.22AKK462 pKa = 9.7GFSPIPSFSTVIPAYY477 pKa = 9.63YY478 pKa = 10.18HH479 pKa = 4.73VTTPVTPKK487 pKa = 10.25TSS489 pKa = 2.93
Molecular weight: 53.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1112 |
489 |
623 |
556.0 |
63.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.554 ± 1.394 | 1.259 ± 0.272 |
5.306 ± 0.749 | 3.957 ± 0.8 |
6.025 ± 0.058 | 4.586 ± 0.324 |
3.867 ± 0.011 | 5.306 ± 0.497 |
3.957 ± 0.926 | 8.004 ± 0.269 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.518 ± 0.039 | 5.665 ± 0.415 |
7.284 ± 0.678 | 3.507 ± 0.359 |
4.676 ± 0.613 | 7.914 ± 1.046 |
6.924 ± 0.774 | 5.306 ± 0.637 |
2.158 ± 0.448 | 4.227 ± 0.967 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
