
Lactobacillus phage ATCCB
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
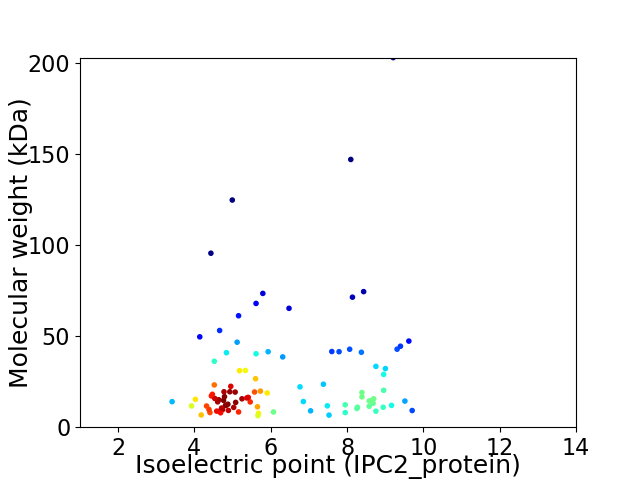
Virtual 2D-PAGE plot for 96 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y5FF61|A0A4Y5FF61_9CAUD Uncharacterized protein OS=Lactobacillus phage ATCCB OX=2510944 GN=ATCCB_0029 PE=4 SV=1
MM1 pKa = 7.58NEE3 pKa = 3.7TEE5 pKa = 3.89EE6 pKa = 4.06QQRR9 pKa = 11.84CKK11 pKa = 10.48YY12 pKa = 10.0CHH14 pKa = 6.73GDD16 pKa = 3.34SEE18 pKa = 6.43LIMDD22 pKa = 4.97DD23 pKa = 3.7GDD25 pKa = 3.69YY26 pKa = 11.52GGIDD30 pKa = 3.57VYY32 pKa = 11.13ISGNILTIDD41 pKa = 4.21YY42 pKa = 10.8INEE45 pKa = 3.83TSINYY50 pKa = 9.61CPMCGRR56 pKa = 11.84KK57 pKa = 9.01LVV59 pKa = 3.43
MM1 pKa = 7.58NEE3 pKa = 3.7TEE5 pKa = 3.89EE6 pKa = 4.06QQRR9 pKa = 11.84CKK11 pKa = 10.48YY12 pKa = 10.0CHH14 pKa = 6.73GDD16 pKa = 3.34SEE18 pKa = 6.43LIMDD22 pKa = 4.97DD23 pKa = 3.7GDD25 pKa = 3.69YY26 pKa = 11.52GGIDD30 pKa = 3.57VYY32 pKa = 11.13ISGNILTIDD41 pKa = 4.21YY42 pKa = 10.8INEE45 pKa = 3.83TSINYY50 pKa = 9.61CPMCGRR56 pKa = 11.84KK57 pKa = 9.01LVV59 pKa = 3.43
Molecular weight: 6.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y5FF74|A0A4Y5FF74_9CAUD PemK-like protein OS=Lactobacillus phage ATCCB OX=2510944 GN=ATCCB_0032 PE=4 SV=1
MM1 pKa = 7.48LKK3 pKa = 10.1KK4 pKa = 10.49LKK6 pKa = 10.75YY7 pKa = 9.86FGLVLVAAFGIFAFSSQAHH26 pKa = 6.9ASTQPVLDD34 pKa = 3.88LSEE37 pKa = 4.0WQGTISHH44 pKa = 6.03SQAIQLRR51 pKa = 11.84QQNPGVILRR60 pKa = 11.84VQYY63 pKa = 10.78GSQYY67 pKa = 10.28ADD69 pKa = 3.0KK70 pKa = 10.88AFAHH74 pKa = 6.49NAMEE78 pKa = 4.61LNSAKK83 pKa = 10.39EE84 pKa = 3.87PFGVYY89 pKa = 9.57SYY91 pKa = 12.04SMYY94 pKa = 10.59TSASDD99 pKa = 3.62ARR101 pKa = 11.84NEE103 pKa = 4.16AKK105 pKa = 9.68TLYY108 pKa = 10.06NRR110 pKa = 11.84SKK112 pKa = 10.73AYY114 pKa = 10.22NPKK117 pKa = 10.01FYY119 pKa = 11.09ANDD122 pKa = 3.44AEE124 pKa = 4.61QYY126 pKa = 6.63TTTSGSYY133 pKa = 10.04RR134 pKa = 11.84SAVKK138 pKa = 9.9AWADD142 pKa = 3.55EE143 pKa = 4.08MHH145 pKa = 7.14KK146 pKa = 9.87LTNKK150 pKa = 9.84PVVLYY155 pKa = 10.49SGSAFYY161 pKa = 10.46RR162 pKa = 11.84SYY164 pKa = 11.0IGTTSGYY171 pKa = 10.69NGFWEE176 pKa = 4.27ANYY179 pKa = 10.66GSRR182 pKa = 11.84RR183 pKa = 11.84WYY185 pKa = 10.89NSALWQYY192 pKa = 9.71TDD194 pKa = 2.81SHH196 pKa = 7.98YY197 pKa = 11.3SIALNKK203 pKa = 10.44SVDD206 pKa = 3.36ASIVMHH212 pKa = 6.13GNWFNTPSKK221 pKa = 11.09SKK223 pKa = 10.29FVYY226 pKa = 10.04GNLRR230 pKa = 11.84VGTLVRR236 pKa = 11.84VEE238 pKa = 4.46KK239 pKa = 10.45NAKK242 pKa = 9.69FYY244 pKa = 9.4STKK247 pKa = 10.75GKK249 pKa = 10.17LDD251 pKa = 3.57PSIAKK256 pKa = 9.96QNLKK260 pKa = 9.93IKK262 pKa = 9.93QIKK265 pKa = 8.67KK266 pKa = 9.47VNVDD270 pKa = 3.24KK271 pKa = 11.4SNEE274 pKa = 3.71IALVYY279 pKa = 10.19HH280 pKa = 6.92GKK282 pKa = 9.5QVIGWVKK289 pKa = 10.6AQDD292 pKa = 3.33LDD294 pKa = 3.92QYY296 pKa = 9.07YY297 pKa = 9.69TSKK300 pKa = 11.14KK301 pKa = 6.78ITKK304 pKa = 9.24VKK306 pKa = 10.5AKK308 pKa = 8.09QTFYY312 pKa = 11.36SYY314 pKa = 11.08VGKK317 pKa = 10.72KK318 pKa = 9.2KK319 pKa = 10.37VKK321 pKa = 10.29KK322 pKa = 9.63NVKK325 pKa = 10.07GDD327 pKa = 3.51VLKK330 pKa = 11.11VSGYY334 pKa = 10.65SFSKK338 pKa = 9.61TNLPRR343 pKa = 11.84FSYY346 pKa = 10.54GKK348 pKa = 9.0YY349 pKa = 8.42TFTANKK355 pKa = 9.69AFVKK359 pKa = 10.04MISTSSKK366 pKa = 8.99NKK368 pKa = 9.64SVKK371 pKa = 9.95KK372 pKa = 10.06SIRR375 pKa = 11.84KK376 pKa = 9.23IYY378 pKa = 7.89TVKK381 pKa = 10.77PGDD384 pKa = 3.85TLSSIAQQYY393 pKa = 7.01GTTVNSLASKK403 pKa = 11.0NNISNPNIIYY413 pKa = 10.47VGEE416 pKa = 4.08KK417 pKa = 10.89LNII420 pKa = 3.78
MM1 pKa = 7.48LKK3 pKa = 10.1KK4 pKa = 10.49LKK6 pKa = 10.75YY7 pKa = 9.86FGLVLVAAFGIFAFSSQAHH26 pKa = 6.9ASTQPVLDD34 pKa = 3.88LSEE37 pKa = 4.0WQGTISHH44 pKa = 6.03SQAIQLRR51 pKa = 11.84QQNPGVILRR60 pKa = 11.84VQYY63 pKa = 10.78GSQYY67 pKa = 10.28ADD69 pKa = 3.0KK70 pKa = 10.88AFAHH74 pKa = 6.49NAMEE78 pKa = 4.61LNSAKK83 pKa = 10.39EE84 pKa = 3.87PFGVYY89 pKa = 9.57SYY91 pKa = 12.04SMYY94 pKa = 10.59TSASDD99 pKa = 3.62ARR101 pKa = 11.84NEE103 pKa = 4.16AKK105 pKa = 9.68TLYY108 pKa = 10.06NRR110 pKa = 11.84SKK112 pKa = 10.73AYY114 pKa = 10.22NPKK117 pKa = 10.01FYY119 pKa = 11.09ANDD122 pKa = 3.44AEE124 pKa = 4.61QYY126 pKa = 6.63TTTSGSYY133 pKa = 10.04RR134 pKa = 11.84SAVKK138 pKa = 9.9AWADD142 pKa = 3.55EE143 pKa = 4.08MHH145 pKa = 7.14KK146 pKa = 9.87LTNKK150 pKa = 9.84PVVLYY155 pKa = 10.49SGSAFYY161 pKa = 10.46RR162 pKa = 11.84SYY164 pKa = 11.0IGTTSGYY171 pKa = 10.69NGFWEE176 pKa = 4.27ANYY179 pKa = 10.66GSRR182 pKa = 11.84RR183 pKa = 11.84WYY185 pKa = 10.89NSALWQYY192 pKa = 9.71TDD194 pKa = 2.81SHH196 pKa = 7.98YY197 pKa = 11.3SIALNKK203 pKa = 10.44SVDD206 pKa = 3.36ASIVMHH212 pKa = 6.13GNWFNTPSKK221 pKa = 11.09SKK223 pKa = 10.29FVYY226 pKa = 10.04GNLRR230 pKa = 11.84VGTLVRR236 pKa = 11.84VEE238 pKa = 4.46KK239 pKa = 10.45NAKK242 pKa = 9.69FYY244 pKa = 9.4STKK247 pKa = 10.75GKK249 pKa = 10.17LDD251 pKa = 3.57PSIAKK256 pKa = 9.96QNLKK260 pKa = 9.93IKK262 pKa = 9.93QIKK265 pKa = 8.67KK266 pKa = 9.47VNVDD270 pKa = 3.24KK271 pKa = 11.4SNEE274 pKa = 3.71IALVYY279 pKa = 10.19HH280 pKa = 6.92GKK282 pKa = 9.5QVIGWVKK289 pKa = 10.6AQDD292 pKa = 3.33LDD294 pKa = 3.92QYY296 pKa = 9.07YY297 pKa = 9.69TSKK300 pKa = 11.14KK301 pKa = 6.78ITKK304 pKa = 9.24VKK306 pKa = 10.5AKK308 pKa = 8.09QTFYY312 pKa = 11.36SYY314 pKa = 11.08VGKK317 pKa = 10.72KK318 pKa = 9.2KK319 pKa = 10.37VKK321 pKa = 10.29KK322 pKa = 9.63NVKK325 pKa = 10.07GDD327 pKa = 3.51VLKK330 pKa = 11.11VSGYY334 pKa = 10.65SFSKK338 pKa = 9.61TNLPRR343 pKa = 11.84FSYY346 pKa = 10.54GKK348 pKa = 9.0YY349 pKa = 8.42TFTANKK355 pKa = 9.69AFVKK359 pKa = 10.04MISTSSKK366 pKa = 8.99NKK368 pKa = 9.64SVKK371 pKa = 9.95KK372 pKa = 10.06SIRR375 pKa = 11.84KK376 pKa = 9.23IYY378 pKa = 7.89TVKK381 pKa = 10.77PGDD384 pKa = 3.85TLSSIAQQYY393 pKa = 7.01GTTVNSLASKK403 pKa = 11.0NNISNPNIIYY413 pKa = 10.47VGEE416 pKa = 4.08KK417 pKa = 10.89LNII420 pKa = 3.78
Molecular weight: 47.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
23815 |
56 |
1859 |
248.1 |
28.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.61 ± 0.491 | 0.869 ± 0.124 |
6.693 ± 0.339 | 5.736 ± 0.297 |
4.212 ± 0.274 | 5.958 ± 0.432 |
1.864 ± 0.12 | 7.819 ± 0.321 |
9.099 ± 0.344 | 7.894 ± 0.242 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.566 ± 0.118 | 7.89 ± 0.314 |
2.734 ± 0.171 | 3.813 ± 0.211 |
3.069 ± 0.136 | 7.239 ± 0.225 |
5.963 ± 0.36 | 5.581 ± 0.164 |
1.012 ± 0.089 | 4.38 ± 0.211 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
