
Bacteroidales bacterium Barb6XT
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; unclassified Bacteroidales
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
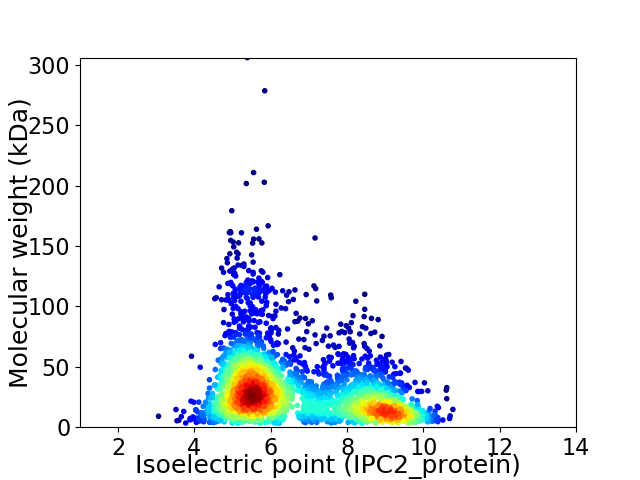
Virtual 2D-PAGE plot for 3068 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A180FAM4|A0A180FAM4_9BACT Uncharacterized protein OS=Bacteroidales bacterium Barb6XT OX=1633202 GN=Barb6XT_00527 PE=4 SV=1
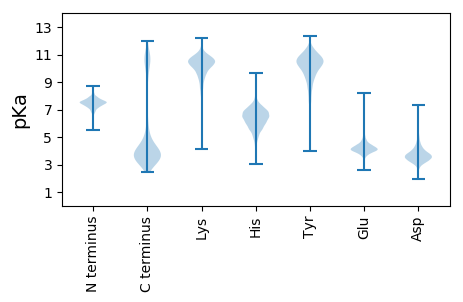
MM1 pKa = 7.36KK2 pKa = 10.63NEE4 pKa = 3.98AQHH7 pKa = 6.82LIDD10 pKa = 4.57RR11 pKa = 11.84FLSARR16 pKa = 11.84NEE18 pKa = 4.01GKK20 pKa = 9.81EE21 pKa = 3.97AYY23 pKa = 9.97FDD25 pKa = 3.96ADD27 pKa = 4.35EE28 pKa = 5.68IDD30 pKa = 5.41DD31 pKa = 4.4LLEE34 pKa = 4.28GCEE37 pKa = 3.98NKK39 pKa = 10.32EE40 pKa = 4.36DD41 pKa = 3.81FDD43 pKa = 5.14LYY45 pKa = 11.37EE46 pKa = 4.26EE47 pKa = 4.32VLALGLKK54 pKa = 9.1LHH56 pKa = 7.15PEE58 pKa = 4.38NISLQTKK65 pKa = 9.58RR66 pKa = 11.84CWQYY70 pKa = 11.12VVEE73 pKa = 4.48EE74 pKa = 4.47NYY76 pKa = 7.72TTALEE81 pKa = 4.59LIEE84 pKa = 4.7QIGATGDD91 pKa = 3.54QEE93 pKa = 4.21IEE95 pKa = 4.05AVQLEE100 pKa = 5.14CYY102 pKa = 10.62LMLDD106 pKa = 3.52MTDD109 pKa = 3.25EE110 pKa = 4.56AEE112 pKa = 4.27ALSEE116 pKa = 4.18KK117 pKa = 9.97MIAEE121 pKa = 4.3GRR123 pKa = 11.84EE124 pKa = 4.03SMDD127 pKa = 3.76LVFEE131 pKa = 4.52YY132 pKa = 10.19IVPILNDD139 pKa = 3.08KK140 pKa = 10.61EE141 pKa = 4.39MYY143 pKa = 10.41SLALEE148 pKa = 4.23YY149 pKa = 11.05VNRR152 pKa = 11.84GLEE155 pKa = 4.33LYY157 pKa = 9.93PDD159 pKa = 3.69SLVLKK164 pKa = 10.56EE165 pKa = 4.13EE166 pKa = 4.38LCFLMEE172 pKa = 3.99VAEE175 pKa = 4.55DD176 pKa = 3.79LEE178 pKa = 4.72GAIDD182 pKa = 4.26LCNAMIDD189 pKa = 3.83EE190 pKa = 5.04DD191 pKa = 4.66PYY193 pKa = 11.43SYY195 pKa = 10.78EE196 pKa = 3.66YY197 pKa = 10.55WFTLGRR203 pKa = 11.84LYY205 pKa = 11.1SLTEE209 pKa = 4.01EE210 pKa = 4.18FEE212 pKa = 4.21KK213 pKa = 11.1AIDD216 pKa = 3.9AFDD219 pKa = 4.04FAQTCEE225 pKa = 4.19EE226 pKa = 4.64PDD228 pKa = 4.47LEE230 pKa = 4.8LKK232 pKa = 10.28LLKK235 pKa = 10.21AYY237 pKa = 10.56CFFMSDD243 pKa = 3.44KK244 pKa = 11.2YY245 pKa = 11.26EE246 pKa = 4.19EE247 pKa = 4.11ALKK250 pKa = 10.78LYY252 pKa = 10.34TEE254 pKa = 4.28LLKK257 pKa = 11.09DD258 pKa = 3.55EE259 pKa = 5.08EE260 pKa = 4.54KK261 pKa = 11.12NEE263 pKa = 3.99EE264 pKa = 4.33DD265 pKa = 5.43GEE267 pKa = 5.0GDD269 pKa = 3.66DD270 pKa = 5.04YY271 pKa = 11.39ILEE274 pKa = 4.22RR275 pKa = 11.84VRR277 pKa = 11.84PLLAEE282 pKa = 4.56CYY284 pKa = 8.12MKK286 pKa = 11.06LFDD289 pKa = 5.07YY290 pKa = 10.03EE291 pKa = 3.82AAYY294 pKa = 10.48LILHH298 pKa = 6.42EE299 pKa = 5.64LIADD303 pKa = 3.83DD304 pKa = 5.42AEE306 pKa = 4.27AAEE309 pKa = 4.5YY310 pKa = 10.28EE311 pKa = 4.34ADD313 pKa = 4.66DD314 pKa = 5.21ADD316 pKa = 4.29TEE318 pKa = 4.93ADD320 pKa = 4.19DD321 pKa = 4.84TDD323 pKa = 4.17TDD325 pKa = 3.72DD326 pKa = 3.52TDD328 pKa = 3.87AARR331 pKa = 11.84KK332 pKa = 9.93DD333 pKa = 3.69YY334 pKa = 11.22SPYY337 pKa = 9.79IYY339 pKa = 10.9YY340 pKa = 7.49MHH342 pKa = 7.46CCAQTDD348 pKa = 3.98RR349 pKa = 11.84KK350 pKa = 10.24QDD352 pKa = 3.48ALDD355 pKa = 4.04ALEE358 pKa = 4.2TAATVFGGSAHH369 pKa = 6.82ILSLMAYY376 pKa = 9.65AYY378 pKa = 9.66MEE380 pKa = 4.27SAEE383 pKa = 4.29NDD385 pKa = 3.58DD386 pKa = 4.75NLDD389 pKa = 3.78VINKK393 pKa = 9.28LFEE396 pKa = 5.87LIDD399 pKa = 4.32DD400 pKa = 5.06PGDD403 pKa = 4.15DD404 pKa = 5.3LEE406 pKa = 4.44QDD408 pKa = 3.57EE409 pKa = 5.76DD410 pKa = 4.01NPSDD414 pKa = 3.63KK415 pKa = 11.24GDD417 pKa = 3.45FSEE420 pKa = 4.88TVGKK424 pKa = 8.97MLEE427 pKa = 4.2YY428 pKa = 10.32FKK430 pKa = 10.7KK431 pKa = 10.29IYY433 pKa = 9.9AANPDD438 pKa = 3.25QPYY441 pKa = 10.56INLYY445 pKa = 10.09LATAYY450 pKa = 10.62LINDD454 pKa = 3.36DD455 pKa = 3.95MEE457 pKa = 4.9NFEE460 pKa = 4.22KK461 pKa = 10.48HH462 pKa = 5.78YY463 pKa = 10.8NLLSAEE469 pKa = 4.34EE470 pKa = 4.08LDD472 pKa = 5.01SYY474 pKa = 10.26AKK476 pKa = 10.26KK477 pKa = 10.91YY478 pKa = 9.94NAGDD482 pKa = 3.44SRR484 pKa = 11.84ALRR487 pKa = 11.84TVAEE491 pKa = 4.2KK492 pKa = 9.26HH493 pKa = 5.24TGLRR497 pKa = 11.84DD498 pKa = 3.24LAEE501 pKa = 4.7EE502 pKa = 4.16YY503 pKa = 10.61LSKK506 pKa = 10.93PEE508 pKa = 3.94NSNN511 pKa = 3.08
MM1 pKa = 7.36KK2 pKa = 10.63NEE4 pKa = 3.98AQHH7 pKa = 6.82LIDD10 pKa = 4.57RR11 pKa = 11.84FLSARR16 pKa = 11.84NEE18 pKa = 4.01GKK20 pKa = 9.81EE21 pKa = 3.97AYY23 pKa = 9.97FDD25 pKa = 3.96ADD27 pKa = 4.35EE28 pKa = 5.68IDD30 pKa = 5.41DD31 pKa = 4.4LLEE34 pKa = 4.28GCEE37 pKa = 3.98NKK39 pKa = 10.32EE40 pKa = 4.36DD41 pKa = 3.81FDD43 pKa = 5.14LYY45 pKa = 11.37EE46 pKa = 4.26EE47 pKa = 4.32VLALGLKK54 pKa = 9.1LHH56 pKa = 7.15PEE58 pKa = 4.38NISLQTKK65 pKa = 9.58RR66 pKa = 11.84CWQYY70 pKa = 11.12VVEE73 pKa = 4.48EE74 pKa = 4.47NYY76 pKa = 7.72TTALEE81 pKa = 4.59LIEE84 pKa = 4.7QIGATGDD91 pKa = 3.54QEE93 pKa = 4.21IEE95 pKa = 4.05AVQLEE100 pKa = 5.14CYY102 pKa = 10.62LMLDD106 pKa = 3.52MTDD109 pKa = 3.25EE110 pKa = 4.56AEE112 pKa = 4.27ALSEE116 pKa = 4.18KK117 pKa = 9.97MIAEE121 pKa = 4.3GRR123 pKa = 11.84EE124 pKa = 4.03SMDD127 pKa = 3.76LVFEE131 pKa = 4.52YY132 pKa = 10.19IVPILNDD139 pKa = 3.08KK140 pKa = 10.61EE141 pKa = 4.39MYY143 pKa = 10.41SLALEE148 pKa = 4.23YY149 pKa = 11.05VNRR152 pKa = 11.84GLEE155 pKa = 4.33LYY157 pKa = 9.93PDD159 pKa = 3.69SLVLKK164 pKa = 10.56EE165 pKa = 4.13EE166 pKa = 4.38LCFLMEE172 pKa = 3.99VAEE175 pKa = 4.55DD176 pKa = 3.79LEE178 pKa = 4.72GAIDD182 pKa = 4.26LCNAMIDD189 pKa = 3.83EE190 pKa = 5.04DD191 pKa = 4.66PYY193 pKa = 11.43SYY195 pKa = 10.78EE196 pKa = 3.66YY197 pKa = 10.55WFTLGRR203 pKa = 11.84LYY205 pKa = 11.1SLTEE209 pKa = 4.01EE210 pKa = 4.18FEE212 pKa = 4.21KK213 pKa = 11.1AIDD216 pKa = 3.9AFDD219 pKa = 4.04FAQTCEE225 pKa = 4.19EE226 pKa = 4.64PDD228 pKa = 4.47LEE230 pKa = 4.8LKK232 pKa = 10.28LLKK235 pKa = 10.21AYY237 pKa = 10.56CFFMSDD243 pKa = 3.44KK244 pKa = 11.2YY245 pKa = 11.26EE246 pKa = 4.19EE247 pKa = 4.11ALKK250 pKa = 10.78LYY252 pKa = 10.34TEE254 pKa = 4.28LLKK257 pKa = 11.09DD258 pKa = 3.55EE259 pKa = 5.08EE260 pKa = 4.54KK261 pKa = 11.12NEE263 pKa = 3.99EE264 pKa = 4.33DD265 pKa = 5.43GEE267 pKa = 5.0GDD269 pKa = 3.66DD270 pKa = 5.04YY271 pKa = 11.39ILEE274 pKa = 4.22RR275 pKa = 11.84VRR277 pKa = 11.84PLLAEE282 pKa = 4.56CYY284 pKa = 8.12MKK286 pKa = 11.06LFDD289 pKa = 5.07YY290 pKa = 10.03EE291 pKa = 3.82AAYY294 pKa = 10.48LILHH298 pKa = 6.42EE299 pKa = 5.64LIADD303 pKa = 3.83DD304 pKa = 5.42AEE306 pKa = 4.27AAEE309 pKa = 4.5YY310 pKa = 10.28EE311 pKa = 4.34ADD313 pKa = 4.66DD314 pKa = 5.21ADD316 pKa = 4.29TEE318 pKa = 4.93ADD320 pKa = 4.19DD321 pKa = 4.84TDD323 pKa = 4.17TDD325 pKa = 3.72DD326 pKa = 3.52TDD328 pKa = 3.87AARR331 pKa = 11.84KK332 pKa = 9.93DD333 pKa = 3.69YY334 pKa = 11.22SPYY337 pKa = 9.79IYY339 pKa = 10.9YY340 pKa = 7.49MHH342 pKa = 7.46CCAQTDD348 pKa = 3.98RR349 pKa = 11.84KK350 pKa = 10.24QDD352 pKa = 3.48ALDD355 pKa = 4.04ALEE358 pKa = 4.2TAATVFGGSAHH369 pKa = 6.82ILSLMAYY376 pKa = 9.65AYY378 pKa = 9.66MEE380 pKa = 4.27SAEE383 pKa = 4.29NDD385 pKa = 3.58DD386 pKa = 4.75NLDD389 pKa = 3.78VINKK393 pKa = 9.28LFEE396 pKa = 5.87LIDD399 pKa = 4.32DD400 pKa = 5.06PGDD403 pKa = 4.15DD404 pKa = 5.3LEE406 pKa = 4.44QDD408 pKa = 3.57EE409 pKa = 5.76DD410 pKa = 4.01NPSDD414 pKa = 3.63KK415 pKa = 11.24GDD417 pKa = 3.45FSEE420 pKa = 4.88TVGKK424 pKa = 8.97MLEE427 pKa = 4.2YY428 pKa = 10.32FKK430 pKa = 10.7KK431 pKa = 10.29IYY433 pKa = 9.9AANPDD438 pKa = 3.25QPYY441 pKa = 10.56INLYY445 pKa = 10.09LATAYY450 pKa = 10.62LINDD454 pKa = 3.36DD455 pKa = 3.95MEE457 pKa = 4.9NFEE460 pKa = 4.22KK461 pKa = 10.48HH462 pKa = 5.78YY463 pKa = 10.8NLLSAEE469 pKa = 4.34EE470 pKa = 4.08LDD472 pKa = 5.01SYY474 pKa = 10.26AKK476 pKa = 10.26KK477 pKa = 10.91YY478 pKa = 9.94NAGDD482 pKa = 3.44SRR484 pKa = 11.84ALRR487 pKa = 11.84TVAEE491 pKa = 4.2KK492 pKa = 9.26HH493 pKa = 5.24TGLRR497 pKa = 11.84DD498 pKa = 3.24LAEE501 pKa = 4.7EE502 pKa = 4.16YY503 pKa = 10.61LSKK506 pKa = 10.93PEE508 pKa = 3.94NSNN511 pKa = 3.08
Molecular weight: 58.81 kDa
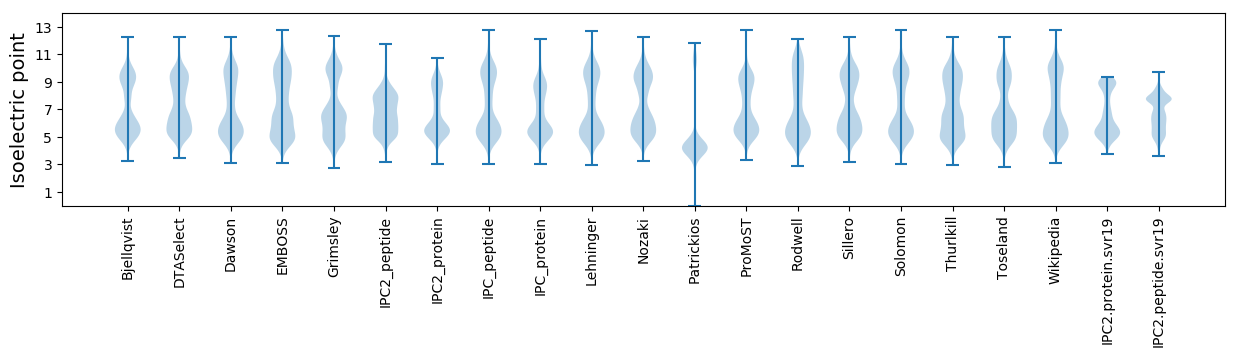
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A180EWT5|A0A180EWT5_9BACT Hef nuclease OS=Bacteroidales bacterium Barb6XT OX=1633202 GN=Barb6XT_02755 PE=4 SV=1
MM1 pKa = 8.09AEE3 pKa = 4.02RR4 pKa = 11.84QRR6 pKa = 11.84IASALNDD13 pKa = 3.51LSLRR17 pKa = 11.84EE18 pKa = 3.96VNMRR22 pKa = 11.84TGHH25 pKa = 7.03LYY27 pKa = 10.59DD28 pKa = 4.25PACIPVPVFAGNPVGRR44 pKa = 11.84CLLAQFLNRR53 pKa = 11.84HH54 pKa = 4.66SRR56 pKa = 11.84QRR58 pKa = 11.84HH59 pKa = 4.14NRR61 pKa = 11.84PPLVTVGLKK70 pKa = 10.72LEE72 pKa = 4.65IIKK75 pKa = 9.51VLPFAGIIHH84 pKa = 6.55CTQQGASAKK93 pKa = 10.28SPVVYY98 pKa = 9.3LYY100 pKa = 10.7CILPVPVQMQLLHH113 pKa = 6.83RR114 pKa = 11.84IVVKK118 pKa = 10.06YY119 pKa = 9.61PPADD123 pKa = 3.32GFHH126 pKa = 7.31PVRR129 pKa = 11.84QRR131 pKa = 11.84IVMPSDD137 pKa = 3.03QMVEE141 pKa = 3.77HH142 pKa = 6.31TVRR145 pKa = 11.84HH146 pKa = 6.04LFHH149 pKa = 6.76IQRR152 pKa = 11.84HH153 pKa = 4.62LLYY156 pKa = 10.0IRR158 pKa = 11.84RR159 pKa = 11.84RR160 pKa = 11.84QHH162 pKa = 5.78QKK164 pKa = 9.62VKK166 pKa = 10.32PIVFAVKK173 pKa = 10.13SYY175 pKa = 9.09PVCFEE180 pKa = 4.0RR181 pKa = 11.84QHH183 pKa = 7.23LSALQIFIGSYY194 pKa = 9.16LGCFRR199 pKa = 11.84KK200 pKa = 10.04HH201 pKa = 4.74IHH203 pKa = 5.79LVQYY207 pKa = 8.19TALPVLKK214 pKa = 10.57VLIRR218 pKa = 11.84VFIFPCFLQCVKK230 pKa = 10.32IFQYY234 pKa = 10.41CFRR237 pKa = 11.84FLFIFRR243 pKa = 11.84KK244 pKa = 10.53GNFHH248 pKa = 5.35TVNRR252 pKa = 11.84RR253 pKa = 11.84SS254 pKa = 3.15
MM1 pKa = 8.09AEE3 pKa = 4.02RR4 pKa = 11.84QRR6 pKa = 11.84IASALNDD13 pKa = 3.51LSLRR17 pKa = 11.84EE18 pKa = 3.96VNMRR22 pKa = 11.84TGHH25 pKa = 7.03LYY27 pKa = 10.59DD28 pKa = 4.25PACIPVPVFAGNPVGRR44 pKa = 11.84CLLAQFLNRR53 pKa = 11.84HH54 pKa = 4.66SRR56 pKa = 11.84QRR58 pKa = 11.84HH59 pKa = 4.14NRR61 pKa = 11.84PPLVTVGLKK70 pKa = 10.72LEE72 pKa = 4.65IIKK75 pKa = 9.51VLPFAGIIHH84 pKa = 6.55CTQQGASAKK93 pKa = 10.28SPVVYY98 pKa = 9.3LYY100 pKa = 10.7CILPVPVQMQLLHH113 pKa = 6.83RR114 pKa = 11.84IVVKK118 pKa = 10.06YY119 pKa = 9.61PPADD123 pKa = 3.32GFHH126 pKa = 7.31PVRR129 pKa = 11.84QRR131 pKa = 11.84IVMPSDD137 pKa = 3.03QMVEE141 pKa = 3.77HH142 pKa = 6.31TVRR145 pKa = 11.84HH146 pKa = 6.04LFHH149 pKa = 6.76IQRR152 pKa = 11.84HH153 pKa = 4.62LLYY156 pKa = 10.0IRR158 pKa = 11.84RR159 pKa = 11.84RR160 pKa = 11.84QHH162 pKa = 5.78QKK164 pKa = 9.62VKK166 pKa = 10.32PIVFAVKK173 pKa = 10.13SYY175 pKa = 9.09PVCFEE180 pKa = 4.0RR181 pKa = 11.84QHH183 pKa = 7.23LSALQIFIGSYY194 pKa = 9.16LGCFRR199 pKa = 11.84KK200 pKa = 10.04HH201 pKa = 4.74IHH203 pKa = 5.79LVQYY207 pKa = 8.19TALPVLKK214 pKa = 10.57VLIRR218 pKa = 11.84VFIFPCFLQCVKK230 pKa = 10.32IFQYY234 pKa = 10.41CFRR237 pKa = 11.84FLFIFRR243 pKa = 11.84KK244 pKa = 10.53GNFHH248 pKa = 5.35TVNRR252 pKa = 11.84RR253 pKa = 11.84SS254 pKa = 3.15
Molecular weight: 29.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
929218 |
29 |
2778 |
302.9 |
33.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.293 ± 0.05 | 1.169 ± 0.022 |
5.495 ± 0.03 | 6.345 ± 0.042 |
4.576 ± 0.033 | 7.34 ± 0.053 |
1.908 ± 0.021 | 6.412 ± 0.045 |
6.113 ± 0.044 | 9.047 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.284 ± 0.024 | 4.7 ± 0.04 |
3.908 ± 0.026 | 3.179 ± 0.028 |
5.12 ± 0.036 | 6.299 ± 0.04 |
5.991 ± 0.045 | 6.672 ± 0.042 |
1.068 ± 0.014 | 4.083 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
