
Changjiang picorna-like virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
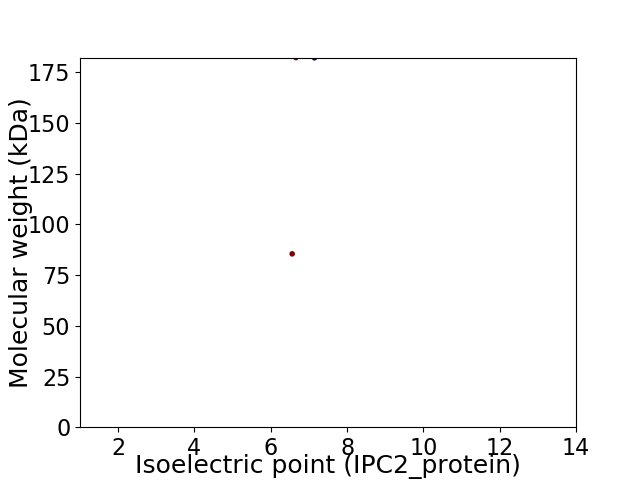
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KNP0|A0A1L3KNP0_9VIRU Uncharacterized protein OS=Changjiang picorna-like virus 6 OX=1922795 PE=4 SV=1
MM1 pKa = 7.17KK2 pKa = 8.13TTAVTMFQNEE12 pKa = 3.9ADD14 pKa = 3.51VCAEE18 pKa = 4.19VNASANPTIVPDD30 pKa = 5.18DD31 pKa = 5.29DD32 pKa = 5.23IGDD35 pKa = 3.84LKK37 pKa = 11.01KK38 pKa = 10.81YY39 pKa = 10.44LSRR42 pKa = 11.84PFPWRR47 pKa = 11.84SGSFTASPGAQEE59 pKa = 4.18TFNISSLGNVTTVFGQGIFDD79 pKa = 3.75KK80 pKa = 10.98MEE82 pKa = 4.12GALGFRR88 pKa = 11.84ATFCFKK94 pKa = 10.87VVVTATPFHH103 pKa = 6.4QGVAAISFQYY113 pKa = 11.33GNISAGNNTRR123 pKa = 11.84GSFYY127 pKa = 11.07YY128 pKa = 10.56LAPNLPHH135 pKa = 6.55VKK137 pKa = 10.25LDD139 pKa = 3.29IAEE142 pKa = 4.22QTSATLKK149 pKa = 10.55VPFISACEE157 pKa = 3.84YY158 pKa = 10.12WPLKK162 pKa = 10.85SEE164 pKa = 4.33AEE166 pKa = 4.04DD167 pKa = 3.32WNVNMGTFAITRR179 pKa = 11.84LTSFRR184 pKa = 11.84LTGTQTAPRR193 pKa = 11.84YY194 pKa = 7.37TVYY197 pKa = 9.65TWLEE201 pKa = 3.95DD202 pKa = 3.67VEE204 pKa = 5.37LVGAYY209 pKa = 9.14PYY211 pKa = 9.2ATKK214 pKa = 10.0TVTFQAGLDD223 pKa = 3.52AEE225 pKa = 4.32LRR227 pKa = 11.84EE228 pKa = 4.41SKK230 pKa = 10.49LVSRR234 pKa = 11.84GLTTVSKK241 pKa = 10.53IAGSLTQVPIIGPIAGHH258 pKa = 5.39TAWFSRR264 pKa = 11.84LLSGAASSFGYY275 pKa = 9.89SRR277 pKa = 11.84PVDD280 pKa = 3.45EE281 pKa = 4.22TVVRR285 pKa = 11.84RR286 pKa = 11.84KK287 pKa = 9.88MIVGYY292 pKa = 10.37AGEE295 pKa = 4.37SHH297 pKa = 7.6VDD299 pKa = 3.48MPSSSFKK306 pKa = 10.1ATPFQTNKK314 pKa = 10.3LAVGKK319 pKa = 10.49LGGNGIDD326 pKa = 3.3EE327 pKa = 4.41MSFEE331 pKa = 5.47HH332 pKa = 7.21ILAKK336 pKa = 10.08PSYY339 pKa = 9.72IYY341 pKa = 10.53RR342 pKa = 11.84KK343 pKa = 9.82QFDD346 pKa = 3.55STGAVGDD353 pKa = 4.29LLYY356 pKa = 10.91CSIVSPSCFWYY367 pKa = 10.13RR368 pKa = 11.84DD369 pKa = 3.52NVGTGNIGLPPNATLTTSAFAPSTIMYY396 pKa = 9.32LGSNFRR402 pKa = 11.84YY403 pKa = 8.32WRR405 pKa = 11.84GKK407 pKa = 9.2FRR409 pKa = 11.84YY410 pKa = 8.99HH411 pKa = 6.41IKK413 pKa = 10.14FSKK416 pKa = 9.65TKK418 pKa = 7.91MHH420 pKa = 7.11GGRR423 pKa = 11.84VLLSFTPSSIFNQNSPITTSMEE445 pKa = 3.92IPDD448 pKa = 4.23SNTAGVQMTGYY459 pKa = 10.92SKK461 pKa = 10.5MFDD464 pKa = 3.4LRR466 pKa = 11.84DD467 pKa = 3.59GSEE470 pKa = 4.03VEE472 pKa = 3.76FDD474 pKa = 4.93IPFTCQSPYY483 pKa = 8.27QTFTGTTGVFAIQIISPLNSPTNASDD509 pKa = 3.29IVDD512 pKa = 3.42MMVSVSALPGFEE524 pKa = 5.0FSAIMPSLIDD534 pKa = 3.49GVAPNGSISDD544 pKa = 3.45AGVYY548 pKa = 9.18KK549 pKa = 10.46QAGGATDD556 pKa = 3.4TSDD559 pKa = 3.01ASMYY563 pKa = 10.59IPGEE567 pKa = 4.09KK568 pKa = 8.67YY569 pKa = 9.2TSVKK573 pKa = 9.59QLMMIPDD580 pKa = 3.25WHH582 pKa = 8.11IFDD585 pKa = 3.9QANATTLTFTLGPWFKK601 pKa = 11.12KK602 pKa = 10.45NYY604 pKa = 9.77LPNTTGTTPIGSTATAAYY622 pKa = 9.2YY623 pKa = 10.35GSKK626 pKa = 9.5SGRR629 pKa = 11.84MQDD632 pKa = 3.62LFAFANGSAEE642 pKa = 4.14YY643 pKa = 9.72TVITDD648 pKa = 4.13KK649 pKa = 11.09PDD651 pKa = 3.25AAATTITVFTSPNDD665 pKa = 3.56TGNTFTAPGSLYY677 pKa = 10.57QKK679 pKa = 10.56GLNEE683 pKa = 4.4FAAHH687 pKa = 5.87TMFEE691 pKa = 4.22NRR693 pKa = 11.84GSFRR697 pKa = 11.84VCVPSFTKK705 pKa = 9.84YY706 pKa = 9.05QRR708 pKa = 11.84VPAALFSGLVGSDD721 pKa = 2.62ATAPSNYY728 pKa = 8.35TVSGVFTAHH737 pKa = 3.73VTAMRR742 pKa = 11.84VRR744 pKa = 11.84NNSLTTTRR752 pKa = 11.84VAFGKK757 pKa = 10.48AAGDD761 pKa = 3.87DD762 pKa = 3.68ATVSQFIGPPLCNFFQATAANSPNPSTLPFF792 pKa = 4.46
MM1 pKa = 7.17KK2 pKa = 8.13TTAVTMFQNEE12 pKa = 3.9ADD14 pKa = 3.51VCAEE18 pKa = 4.19VNASANPTIVPDD30 pKa = 5.18DD31 pKa = 5.29DD32 pKa = 5.23IGDD35 pKa = 3.84LKK37 pKa = 11.01KK38 pKa = 10.81YY39 pKa = 10.44LSRR42 pKa = 11.84PFPWRR47 pKa = 11.84SGSFTASPGAQEE59 pKa = 4.18TFNISSLGNVTTVFGQGIFDD79 pKa = 3.75KK80 pKa = 10.98MEE82 pKa = 4.12GALGFRR88 pKa = 11.84ATFCFKK94 pKa = 10.87VVVTATPFHH103 pKa = 6.4QGVAAISFQYY113 pKa = 11.33GNISAGNNTRR123 pKa = 11.84GSFYY127 pKa = 11.07YY128 pKa = 10.56LAPNLPHH135 pKa = 6.55VKK137 pKa = 10.25LDD139 pKa = 3.29IAEE142 pKa = 4.22QTSATLKK149 pKa = 10.55VPFISACEE157 pKa = 3.84YY158 pKa = 10.12WPLKK162 pKa = 10.85SEE164 pKa = 4.33AEE166 pKa = 4.04DD167 pKa = 3.32WNVNMGTFAITRR179 pKa = 11.84LTSFRR184 pKa = 11.84LTGTQTAPRR193 pKa = 11.84YY194 pKa = 7.37TVYY197 pKa = 9.65TWLEE201 pKa = 3.95DD202 pKa = 3.67VEE204 pKa = 5.37LVGAYY209 pKa = 9.14PYY211 pKa = 9.2ATKK214 pKa = 10.0TVTFQAGLDD223 pKa = 3.52AEE225 pKa = 4.32LRR227 pKa = 11.84EE228 pKa = 4.41SKK230 pKa = 10.49LVSRR234 pKa = 11.84GLTTVSKK241 pKa = 10.53IAGSLTQVPIIGPIAGHH258 pKa = 5.39TAWFSRR264 pKa = 11.84LLSGAASSFGYY275 pKa = 9.89SRR277 pKa = 11.84PVDD280 pKa = 3.45EE281 pKa = 4.22TVVRR285 pKa = 11.84RR286 pKa = 11.84KK287 pKa = 9.88MIVGYY292 pKa = 10.37AGEE295 pKa = 4.37SHH297 pKa = 7.6VDD299 pKa = 3.48MPSSSFKK306 pKa = 10.1ATPFQTNKK314 pKa = 10.3LAVGKK319 pKa = 10.49LGGNGIDD326 pKa = 3.3EE327 pKa = 4.41MSFEE331 pKa = 5.47HH332 pKa = 7.21ILAKK336 pKa = 10.08PSYY339 pKa = 9.72IYY341 pKa = 10.53RR342 pKa = 11.84KK343 pKa = 9.82QFDD346 pKa = 3.55STGAVGDD353 pKa = 4.29LLYY356 pKa = 10.91CSIVSPSCFWYY367 pKa = 10.13RR368 pKa = 11.84DD369 pKa = 3.52NVGTGNIGLPPNATLTTSAFAPSTIMYY396 pKa = 9.32LGSNFRR402 pKa = 11.84YY403 pKa = 8.32WRR405 pKa = 11.84GKK407 pKa = 9.2FRR409 pKa = 11.84YY410 pKa = 8.99HH411 pKa = 6.41IKK413 pKa = 10.14FSKK416 pKa = 9.65TKK418 pKa = 7.91MHH420 pKa = 7.11GGRR423 pKa = 11.84VLLSFTPSSIFNQNSPITTSMEE445 pKa = 3.92IPDD448 pKa = 4.23SNTAGVQMTGYY459 pKa = 10.92SKK461 pKa = 10.5MFDD464 pKa = 3.4LRR466 pKa = 11.84DD467 pKa = 3.59GSEE470 pKa = 4.03VEE472 pKa = 3.76FDD474 pKa = 4.93IPFTCQSPYY483 pKa = 8.27QTFTGTTGVFAIQIISPLNSPTNASDD509 pKa = 3.29IVDD512 pKa = 3.42MMVSVSALPGFEE524 pKa = 5.0FSAIMPSLIDD534 pKa = 3.49GVAPNGSISDD544 pKa = 3.45AGVYY548 pKa = 9.18KK549 pKa = 10.46QAGGATDD556 pKa = 3.4TSDD559 pKa = 3.01ASMYY563 pKa = 10.59IPGEE567 pKa = 4.09KK568 pKa = 8.67YY569 pKa = 9.2TSVKK573 pKa = 9.59QLMMIPDD580 pKa = 3.25WHH582 pKa = 8.11IFDD585 pKa = 3.9QANATTLTFTLGPWFKK601 pKa = 11.12KK602 pKa = 10.45NYY604 pKa = 9.77LPNTTGTTPIGSTATAAYY622 pKa = 9.2YY623 pKa = 10.35GSKK626 pKa = 9.5SGRR629 pKa = 11.84MQDD632 pKa = 3.62LFAFANGSAEE642 pKa = 4.14YY643 pKa = 9.72TVITDD648 pKa = 4.13KK649 pKa = 11.09PDD651 pKa = 3.25AAATTITVFTSPNDD665 pKa = 3.56TGNTFTAPGSLYY677 pKa = 10.57QKK679 pKa = 10.56GLNEE683 pKa = 4.4FAAHH687 pKa = 5.87TMFEE691 pKa = 4.22NRR693 pKa = 11.84GSFRR697 pKa = 11.84VCVPSFTKK705 pKa = 9.84YY706 pKa = 9.05QRR708 pKa = 11.84VPAALFSGLVGSDD721 pKa = 2.62ATAPSNYY728 pKa = 8.35TVSGVFTAHH737 pKa = 3.73VTAMRR742 pKa = 11.84VRR744 pKa = 11.84NNSLTTTRR752 pKa = 11.84VAFGKK757 pKa = 10.48AAGDD761 pKa = 3.87DD762 pKa = 3.68ATVSQFIGPPLCNFFQATAANSPNPSTLPFF792 pKa = 4.46
Molecular weight: 85.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KNP0|A0A1L3KNP0_9VIRU Uncharacterized protein OS=Changjiang picorna-like virus 6 OX=1922795 PE=4 SV=1
MM1 pKa = 7.61HH2 pKa = 7.59IACQFFLTLGYY13 pKa = 9.74KK14 pKa = 10.11QSTHH18 pKa = 5.86QIQRR22 pKa = 11.84TMNAFNKK29 pKa = 8.59TNAVSTLVRR38 pKa = 11.84RR39 pKa = 11.84YY40 pKa = 7.73TDD42 pKa = 2.88KK43 pKa = 11.15SLQVGDD49 pKa = 3.31NKK51 pKa = 9.86QRR53 pKa = 11.84RR54 pKa = 11.84KK55 pKa = 10.54FEE57 pKa = 4.12MLTKK61 pKa = 9.86QDD63 pKa = 3.19KK64 pKa = 10.23RR65 pKa = 11.84DD66 pKa = 3.18WRR68 pKa = 11.84EE69 pKa = 3.12ALQRR73 pKa = 11.84HH74 pKa = 5.48IAALRR79 pKa = 11.84SRR81 pKa = 11.84AGSRR85 pKa = 11.84PLRR88 pKa = 11.84RR89 pKa = 11.84AFQSKK94 pKa = 9.98PKK96 pKa = 10.23GKK98 pKa = 9.42NAHH101 pKa = 6.39KK102 pKa = 10.16INRR105 pKa = 11.84ISRR108 pKa = 11.84YY109 pKa = 8.19EE110 pKa = 3.76EE111 pKa = 4.01SLASMVMGSDD121 pKa = 4.02GYY123 pKa = 10.38VEE125 pKa = 4.14KK126 pKa = 10.78QSYY129 pKa = 10.7ASAATIGVAAALSTTGAYY147 pKa = 10.66YY148 pKa = 9.64MYY150 pKa = 10.95KK151 pKa = 10.01RR152 pKa = 11.84LSKK155 pKa = 10.28MADD158 pKa = 3.06KK159 pKa = 10.76VVSVSDD165 pKa = 4.01SIGAQVNDD173 pKa = 5.57GIAKK177 pKa = 9.15FRR179 pKa = 11.84QYY181 pKa = 11.08FSDD184 pKa = 5.7FIDD187 pKa = 3.27SLKK190 pKa = 10.8RR191 pKa = 11.84VGSWLAKK198 pKa = 8.78FTICAAGLWFVNKK211 pKa = 9.71YY212 pKa = 9.6VNAPALSVVITSMVLASVPEE232 pKa = 3.85ASNLFGIQKK241 pKa = 9.94QSFDD245 pKa = 3.67GSCNLIALLFTLLAPTIGGKK265 pKa = 9.99YY266 pKa = 10.0GFMVNTFMRR275 pKa = 11.84TVGAFPKK282 pKa = 10.17FSEE285 pKa = 4.29GLPVFMDD292 pKa = 4.21KK293 pKa = 10.85IMDD296 pKa = 3.94VVEE299 pKa = 5.02KK300 pKa = 10.79AIDD303 pKa = 3.84WILKK307 pKa = 7.8RR308 pKa = 11.84TTGGSFSFEE317 pKa = 3.62RR318 pKa = 11.84KK319 pKa = 8.88MDD321 pKa = 3.66LTTKK325 pKa = 8.85WRR327 pKa = 11.84NEE329 pKa = 3.62VLAMCNEE336 pKa = 4.22VDD338 pKa = 3.81TQPKK342 pKa = 9.42VSLEE346 pKa = 4.3TVHH349 pKa = 7.08AMQKK353 pKa = 10.05KK354 pKa = 7.94VQEE357 pKa = 5.01GYY359 pKa = 11.69GLMQLMTHH367 pKa = 6.12KK368 pKa = 10.41QSKK371 pKa = 10.74DD372 pKa = 3.03EE373 pKa = 3.9VARR376 pKa = 11.84YY377 pKa = 9.34IDD379 pKa = 3.64RR380 pKa = 11.84LNSRR384 pKa = 11.84LAPHH388 pKa = 7.31LGTLASEE395 pKa = 4.23NNMRR399 pKa = 11.84VMPYY403 pKa = 10.36CAMLGGGSGIGKK415 pKa = 7.79TSVVQVYY422 pKa = 10.76ASMILVLAGEE432 pKa = 4.59VKK434 pKa = 10.59ASEE437 pKa = 4.26VLQNLWQKK445 pKa = 10.9GISEE449 pKa = 4.37YY450 pKa = 10.4WNGYY454 pKa = 8.3LGQRR458 pKa = 11.84AIIKK462 pKa = 9.69DD463 pKa = 3.25DD464 pKa = 3.63CFQVRR469 pKa = 11.84GVAGAQDD476 pKa = 3.9SEE478 pKa = 4.27AMEE481 pKa = 5.21LIRR484 pKa = 11.84AVGNWACPLNYY495 pKa = 10.21ADD497 pKa = 4.28VDD499 pKa = 4.24SKK501 pKa = 11.32GRR503 pKa = 11.84YY504 pKa = 8.12YY505 pKa = 11.16LDD507 pKa = 3.06VALIVGTTNAANIRR521 pKa = 11.84ADD523 pKa = 3.42WEE525 pKa = 4.17PFITCPEE532 pKa = 3.82ALVRR536 pKa = 11.84RR537 pKa = 11.84FQGAYY542 pKa = 8.11WLEE545 pKa = 3.99LNKK548 pKa = 10.51EE549 pKa = 4.32YY550 pKa = 10.83EE551 pKa = 4.06NDD553 pKa = 2.93MGRR556 pKa = 11.84FDD558 pKa = 3.88FEE560 pKa = 4.22KK561 pKa = 10.46VSRR564 pKa = 11.84VYY566 pKa = 10.95ASRR569 pKa = 11.84LSSFAARR576 pKa = 11.84VASEE580 pKa = 4.28PTWKK584 pKa = 8.79PTEE587 pKa = 4.62DD588 pKa = 3.77DD589 pKa = 4.08VLDD592 pKa = 4.37LFPWDD597 pKa = 2.92AWTVHH602 pKa = 5.24MHH604 pKa = 6.91DD605 pKa = 5.1FSNSNPLNGPVLPGGMKK622 pKa = 10.0QAVKK626 pKa = 10.54DD627 pKa = 3.53AANSIKK633 pKa = 10.3ARR635 pKa = 11.84KK636 pKa = 8.04AAHH639 pKa = 7.16RR640 pKa = 11.84DD641 pKa = 3.25TVQNITEE648 pKa = 4.13HH649 pKa = 6.48LKK651 pKa = 10.3VAEE654 pKa = 4.15SVIDD658 pKa = 3.65KK659 pKa = 10.83LFVDD663 pKa = 4.47KK664 pKa = 11.03QAGATIVGEE673 pKa = 4.47ISATTSDD680 pKa = 3.27NFEE683 pKa = 4.17RR684 pKa = 11.84DD685 pKa = 3.01LRR687 pKa = 11.84ASYY690 pKa = 9.85PGQSIQYY697 pKa = 7.98STVEE701 pKa = 4.01VKK703 pKa = 10.93GLLTAGTDD711 pKa = 3.84GEE713 pKa = 4.96PILLDD718 pKa = 3.55LEE720 pKa = 4.73RR721 pKa = 11.84RR722 pKa = 11.84DD723 pKa = 3.52ATFCEE728 pKa = 4.49TVKK731 pKa = 11.12ANLVAWANRR740 pKa = 11.84LVSYY744 pKa = 11.1VGDD747 pKa = 3.94TLDD750 pKa = 3.42KK751 pKa = 10.76TLGPSVGEE759 pKa = 4.15FNGEE763 pKa = 3.6PTLNVARR770 pKa = 11.84VAVEE774 pKa = 3.89GALCGLALTCVFKK787 pKa = 11.01LIHH790 pKa = 6.19GAVGMLWAMVEE801 pKa = 4.24ALFKK805 pKa = 10.98AVGINTQSNAPPAQSKK821 pKa = 11.11DD822 pKa = 3.09KK823 pKa = 10.81GLKK826 pKa = 9.82KK827 pKa = 10.71FEE829 pKa = 4.13FPKK832 pKa = 10.79VSLQLGAPPQEE843 pKa = 4.05HH844 pKa = 5.6VHH846 pKa = 6.61DD847 pKa = 3.75NVYY850 pKa = 11.32NNMYY854 pKa = 10.46AIGYY858 pKa = 9.21DD859 pKa = 3.48DD860 pKa = 4.96GGVYY864 pKa = 9.38MPVGNIIGIGGQVFVMPAHH883 pKa = 6.65FDD885 pKa = 3.61DD886 pKa = 4.98YY887 pKa = 11.26LAKK890 pKa = 10.31HH891 pKa = 6.27ANADD895 pKa = 3.48ADD897 pKa = 4.02VVLIMCSNVHH907 pKa = 6.54MITRR911 pKa = 11.84IPLKK915 pKa = 9.97LFKK918 pKa = 10.42SFRR921 pKa = 11.84RR922 pKa = 11.84ARR924 pKa = 11.84FEE926 pKa = 4.01STTDD930 pKa = 3.07MVGISFEE937 pKa = 4.12KK938 pKa = 9.73YY939 pKa = 10.04APIRR943 pKa = 11.84QHH945 pKa = 6.15RR946 pKa = 11.84VIVGYY951 pKa = 8.06FLKK954 pKa = 9.8EE955 pKa = 4.13TEE957 pKa = 3.83ISNILRR963 pKa = 11.84GTNVAVRR970 pKa = 11.84LDD972 pKa = 3.01IGRR975 pKa = 11.84RR976 pKa = 11.84RR977 pKa = 11.84KK978 pKa = 10.25DD979 pKa = 3.19KK980 pKa = 10.89EE981 pKa = 3.9IVRR984 pKa = 11.84TTLMSHH990 pKa = 6.48RR991 pKa = 11.84SEE993 pKa = 4.27YY994 pKa = 11.11VPMVSANDD1002 pKa = 4.11GSKK1005 pKa = 8.99LTSLIRR1011 pKa = 11.84YY1012 pKa = 7.56DD1013 pKa = 4.01MPTMSGDD1020 pKa = 3.34CGAPLMLSEE1029 pKa = 3.58NRR1031 pKa = 11.84YY1032 pKa = 9.47YY1033 pKa = 10.88GGRR1036 pKa = 11.84CILGLHH1042 pKa = 5.69VAGKK1046 pKa = 5.21TTLMSRR1052 pKa = 11.84EE1053 pKa = 4.52GYY1055 pKa = 9.96SVIASQEE1062 pKa = 3.97CMRR1065 pKa = 11.84EE1066 pKa = 3.13IWLRR1070 pKa = 11.84LGPSEE1075 pKa = 4.14EE1076 pKa = 4.4VEE1078 pKa = 3.88VDD1080 pKa = 3.7TVHH1083 pKa = 6.93SQMRR1087 pKa = 11.84QVTNEE1092 pKa = 3.69EE1093 pKa = 4.47FVTLEE1098 pKa = 3.95AGLVEE1103 pKa = 4.43KK1104 pKa = 10.8GIIGGSMSYY1113 pKa = 10.32LGPLNEE1119 pKa = 4.68PVNLAPKK1126 pKa = 8.76TALIPSPMHH1135 pKa = 6.4ADD1137 pKa = 3.65EE1138 pKa = 5.31PFGPCPVAPAILYY1151 pKa = 8.31PVVKK1155 pKa = 10.27DD1156 pKa = 3.53GLRR1159 pKa = 11.84VYY1161 pKa = 10.78PMARR1165 pKa = 11.84AVEE1168 pKa = 5.14AYY1170 pKa = 10.26QSDD1173 pKa = 4.65VVVKK1177 pKa = 10.34APQVLDD1183 pKa = 3.35IAAEE1187 pKa = 4.3VAFKK1191 pKa = 10.76PLMKK1195 pKa = 9.63VTQDD1199 pKa = 3.31FPRR1202 pKa = 11.84DD1203 pKa = 3.48VLSFEE1208 pKa = 4.4EE1209 pKa = 4.63AVVPPEE1215 pKa = 3.76GWKK1218 pKa = 10.54LKK1220 pKa = 9.63PLNRR1224 pKa = 11.84KK1225 pKa = 9.06SSAGYY1230 pKa = 9.79KK1231 pKa = 8.41YY1232 pKa = 10.37RR1233 pKa = 11.84SYY1235 pKa = 9.99VTAAKK1240 pKa = 9.18PGKK1243 pKa = 9.81VAFLGKK1249 pKa = 10.45EE1250 pKa = 3.72GDD1252 pKa = 3.58VDD1254 pKa = 4.59FSRR1257 pKa = 11.84PEE1259 pKa = 3.63LDD1261 pKa = 3.01IVRR1264 pKa = 11.84KK1265 pKa = 9.8DD1266 pKa = 3.24VTSIISHH1273 pKa = 6.69AKK1275 pKa = 8.92RR1276 pKa = 11.84GVRR1279 pKa = 11.84LPHH1282 pKa = 6.22YY1283 pKa = 9.06CTDD1286 pKa = 3.81FLKK1289 pKa = 11.08DD1290 pKa = 3.39EE1291 pKa = 4.55LRR1293 pKa = 11.84PLEE1296 pKa = 4.18KK1297 pKa = 10.74VEE1299 pKa = 4.0AVKK1302 pKa = 10.05TRR1304 pKa = 11.84MISGTEE1310 pKa = 3.67LDD1312 pKa = 3.44YY1313 pKa = 11.28TIAVRR1318 pKa = 11.84MYY1320 pKa = 9.9FGAFNAAMLATPVVNGMAPGINHH1343 pKa = 4.68YY1344 pKa = 6.97TQWGEE1349 pKa = 3.57LATRR1353 pKa = 11.84LISKK1357 pKa = 10.17GGAVFDD1363 pKa = 4.49GDD1365 pKa = 3.98FSRR1368 pKa = 11.84FDD1370 pKa = 3.43ASEE1373 pKa = 4.1QPWVHH1378 pKa = 5.71MKK1380 pKa = 10.02ILEE1383 pKa = 5.15VINQWYY1389 pKa = 9.62AMKK1392 pKa = 10.7GGTEE1396 pKa = 3.58EE1397 pKa = 4.44DD1398 pKa = 3.47DD1399 pKa = 3.57RR1400 pKa = 11.84VRR1402 pKa = 11.84TILWEE1407 pKa = 4.06DD1408 pKa = 3.91VIHH1411 pKa = 6.23SVHH1414 pKa = 5.73ITGDD1418 pKa = 3.74SSSHH1422 pKa = 5.24GQLVQWHH1429 pKa = 6.6KK1430 pKa = 10.87SLPSGHH1436 pKa = 7.47PLTTVINSMYY1446 pKa = 10.77SLLALTTCYY1455 pKa = 10.32IHH1457 pKa = 6.57LTGDD1461 pKa = 3.42SRR1463 pKa = 11.84DD1464 pKa = 3.28MWEE1467 pKa = 4.15HH1468 pKa = 5.31VFINTFGDD1476 pKa = 3.83DD1477 pKa = 3.29NVAGVDD1483 pKa = 3.51EE1484 pKa = 4.45SVRR1487 pKa = 11.84DD1488 pKa = 3.56VFNQVTVASAMSEE1501 pKa = 4.19LFNLTYY1507 pKa = 9.81TAGAKK1512 pKa = 9.79DD1513 pKa = 3.97GKK1515 pKa = 10.15LVPYY1519 pKa = 9.77TDD1521 pKa = 3.5IYY1523 pKa = 11.18NITFLKK1529 pKa = 10.47RR1530 pKa = 11.84SFLRR1534 pKa = 11.84DD1535 pKa = 3.38EE1536 pKa = 4.26EE1537 pKa = 4.35TSDD1540 pKa = 3.74IIGSAPCMDD1549 pKa = 3.42WVGPLAKK1556 pKa = 10.19EE1557 pKa = 4.1SFLYY1561 pKa = 9.05TPYY1564 pKa = 10.92YY1565 pKa = 9.57YY1566 pKa = 10.41RR1567 pKa = 11.84NKK1569 pKa = 10.08KK1570 pKa = 10.21DD1571 pKa = 3.29PRR1573 pKa = 11.84KK1574 pKa = 10.49DD1575 pKa = 3.0ITDD1578 pKa = 3.43NCDD1581 pKa = 2.96ILLGEE1586 pKa = 4.69LALHH1590 pKa = 6.65PKK1592 pKa = 10.46SMWDD1596 pKa = 3.32EE1597 pKa = 3.96YY1598 pKa = 11.36FPLLKK1603 pKa = 10.03QWCVKK1608 pKa = 10.7NDD1610 pKa = 3.11IEE1612 pKa = 4.53LTFEE1616 pKa = 3.97SRR1618 pKa = 11.84SAARR1622 pKa = 11.84AYY1624 pKa = 8.36ITTRR1628 pKa = 11.84FDD1630 pKa = 2.83VWFF1633 pKa = 4.48
MM1 pKa = 7.61HH2 pKa = 7.59IACQFFLTLGYY13 pKa = 9.74KK14 pKa = 10.11QSTHH18 pKa = 5.86QIQRR22 pKa = 11.84TMNAFNKK29 pKa = 8.59TNAVSTLVRR38 pKa = 11.84RR39 pKa = 11.84YY40 pKa = 7.73TDD42 pKa = 2.88KK43 pKa = 11.15SLQVGDD49 pKa = 3.31NKK51 pKa = 9.86QRR53 pKa = 11.84RR54 pKa = 11.84KK55 pKa = 10.54FEE57 pKa = 4.12MLTKK61 pKa = 9.86QDD63 pKa = 3.19KK64 pKa = 10.23RR65 pKa = 11.84DD66 pKa = 3.18WRR68 pKa = 11.84EE69 pKa = 3.12ALQRR73 pKa = 11.84HH74 pKa = 5.48IAALRR79 pKa = 11.84SRR81 pKa = 11.84AGSRR85 pKa = 11.84PLRR88 pKa = 11.84RR89 pKa = 11.84AFQSKK94 pKa = 9.98PKK96 pKa = 10.23GKK98 pKa = 9.42NAHH101 pKa = 6.39KK102 pKa = 10.16INRR105 pKa = 11.84ISRR108 pKa = 11.84YY109 pKa = 8.19EE110 pKa = 3.76EE111 pKa = 4.01SLASMVMGSDD121 pKa = 4.02GYY123 pKa = 10.38VEE125 pKa = 4.14KK126 pKa = 10.78QSYY129 pKa = 10.7ASAATIGVAAALSTTGAYY147 pKa = 10.66YY148 pKa = 9.64MYY150 pKa = 10.95KK151 pKa = 10.01RR152 pKa = 11.84LSKK155 pKa = 10.28MADD158 pKa = 3.06KK159 pKa = 10.76VVSVSDD165 pKa = 4.01SIGAQVNDD173 pKa = 5.57GIAKK177 pKa = 9.15FRR179 pKa = 11.84QYY181 pKa = 11.08FSDD184 pKa = 5.7FIDD187 pKa = 3.27SLKK190 pKa = 10.8RR191 pKa = 11.84VGSWLAKK198 pKa = 8.78FTICAAGLWFVNKK211 pKa = 9.71YY212 pKa = 9.6VNAPALSVVITSMVLASVPEE232 pKa = 3.85ASNLFGIQKK241 pKa = 9.94QSFDD245 pKa = 3.67GSCNLIALLFTLLAPTIGGKK265 pKa = 9.99YY266 pKa = 10.0GFMVNTFMRR275 pKa = 11.84TVGAFPKK282 pKa = 10.17FSEE285 pKa = 4.29GLPVFMDD292 pKa = 4.21KK293 pKa = 10.85IMDD296 pKa = 3.94VVEE299 pKa = 5.02KK300 pKa = 10.79AIDD303 pKa = 3.84WILKK307 pKa = 7.8RR308 pKa = 11.84TTGGSFSFEE317 pKa = 3.62RR318 pKa = 11.84KK319 pKa = 8.88MDD321 pKa = 3.66LTTKK325 pKa = 8.85WRR327 pKa = 11.84NEE329 pKa = 3.62VLAMCNEE336 pKa = 4.22VDD338 pKa = 3.81TQPKK342 pKa = 9.42VSLEE346 pKa = 4.3TVHH349 pKa = 7.08AMQKK353 pKa = 10.05KK354 pKa = 7.94VQEE357 pKa = 5.01GYY359 pKa = 11.69GLMQLMTHH367 pKa = 6.12KK368 pKa = 10.41QSKK371 pKa = 10.74DD372 pKa = 3.03EE373 pKa = 3.9VARR376 pKa = 11.84YY377 pKa = 9.34IDD379 pKa = 3.64RR380 pKa = 11.84LNSRR384 pKa = 11.84LAPHH388 pKa = 7.31LGTLASEE395 pKa = 4.23NNMRR399 pKa = 11.84VMPYY403 pKa = 10.36CAMLGGGSGIGKK415 pKa = 7.79TSVVQVYY422 pKa = 10.76ASMILVLAGEE432 pKa = 4.59VKK434 pKa = 10.59ASEE437 pKa = 4.26VLQNLWQKK445 pKa = 10.9GISEE449 pKa = 4.37YY450 pKa = 10.4WNGYY454 pKa = 8.3LGQRR458 pKa = 11.84AIIKK462 pKa = 9.69DD463 pKa = 3.25DD464 pKa = 3.63CFQVRR469 pKa = 11.84GVAGAQDD476 pKa = 3.9SEE478 pKa = 4.27AMEE481 pKa = 5.21LIRR484 pKa = 11.84AVGNWACPLNYY495 pKa = 10.21ADD497 pKa = 4.28VDD499 pKa = 4.24SKK501 pKa = 11.32GRR503 pKa = 11.84YY504 pKa = 8.12YY505 pKa = 11.16LDD507 pKa = 3.06VALIVGTTNAANIRR521 pKa = 11.84ADD523 pKa = 3.42WEE525 pKa = 4.17PFITCPEE532 pKa = 3.82ALVRR536 pKa = 11.84RR537 pKa = 11.84FQGAYY542 pKa = 8.11WLEE545 pKa = 3.99LNKK548 pKa = 10.51EE549 pKa = 4.32YY550 pKa = 10.83EE551 pKa = 4.06NDD553 pKa = 2.93MGRR556 pKa = 11.84FDD558 pKa = 3.88FEE560 pKa = 4.22KK561 pKa = 10.46VSRR564 pKa = 11.84VYY566 pKa = 10.95ASRR569 pKa = 11.84LSSFAARR576 pKa = 11.84VASEE580 pKa = 4.28PTWKK584 pKa = 8.79PTEE587 pKa = 4.62DD588 pKa = 3.77DD589 pKa = 4.08VLDD592 pKa = 4.37LFPWDD597 pKa = 2.92AWTVHH602 pKa = 5.24MHH604 pKa = 6.91DD605 pKa = 5.1FSNSNPLNGPVLPGGMKK622 pKa = 10.0QAVKK626 pKa = 10.54DD627 pKa = 3.53AANSIKK633 pKa = 10.3ARR635 pKa = 11.84KK636 pKa = 8.04AAHH639 pKa = 7.16RR640 pKa = 11.84DD641 pKa = 3.25TVQNITEE648 pKa = 4.13HH649 pKa = 6.48LKK651 pKa = 10.3VAEE654 pKa = 4.15SVIDD658 pKa = 3.65KK659 pKa = 10.83LFVDD663 pKa = 4.47KK664 pKa = 11.03QAGATIVGEE673 pKa = 4.47ISATTSDD680 pKa = 3.27NFEE683 pKa = 4.17RR684 pKa = 11.84DD685 pKa = 3.01LRR687 pKa = 11.84ASYY690 pKa = 9.85PGQSIQYY697 pKa = 7.98STVEE701 pKa = 4.01VKK703 pKa = 10.93GLLTAGTDD711 pKa = 3.84GEE713 pKa = 4.96PILLDD718 pKa = 3.55LEE720 pKa = 4.73RR721 pKa = 11.84RR722 pKa = 11.84DD723 pKa = 3.52ATFCEE728 pKa = 4.49TVKK731 pKa = 11.12ANLVAWANRR740 pKa = 11.84LVSYY744 pKa = 11.1VGDD747 pKa = 3.94TLDD750 pKa = 3.42KK751 pKa = 10.76TLGPSVGEE759 pKa = 4.15FNGEE763 pKa = 3.6PTLNVARR770 pKa = 11.84VAVEE774 pKa = 3.89GALCGLALTCVFKK787 pKa = 11.01LIHH790 pKa = 6.19GAVGMLWAMVEE801 pKa = 4.24ALFKK805 pKa = 10.98AVGINTQSNAPPAQSKK821 pKa = 11.11DD822 pKa = 3.09KK823 pKa = 10.81GLKK826 pKa = 9.82KK827 pKa = 10.71FEE829 pKa = 4.13FPKK832 pKa = 10.79VSLQLGAPPQEE843 pKa = 4.05HH844 pKa = 5.6VHH846 pKa = 6.61DD847 pKa = 3.75NVYY850 pKa = 11.32NNMYY854 pKa = 10.46AIGYY858 pKa = 9.21DD859 pKa = 3.48DD860 pKa = 4.96GGVYY864 pKa = 9.38MPVGNIIGIGGQVFVMPAHH883 pKa = 6.65FDD885 pKa = 3.61DD886 pKa = 4.98YY887 pKa = 11.26LAKK890 pKa = 10.31HH891 pKa = 6.27ANADD895 pKa = 3.48ADD897 pKa = 4.02VVLIMCSNVHH907 pKa = 6.54MITRR911 pKa = 11.84IPLKK915 pKa = 9.97LFKK918 pKa = 10.42SFRR921 pKa = 11.84RR922 pKa = 11.84ARR924 pKa = 11.84FEE926 pKa = 4.01STTDD930 pKa = 3.07MVGISFEE937 pKa = 4.12KK938 pKa = 9.73YY939 pKa = 10.04APIRR943 pKa = 11.84QHH945 pKa = 6.15RR946 pKa = 11.84VIVGYY951 pKa = 8.06FLKK954 pKa = 9.8EE955 pKa = 4.13TEE957 pKa = 3.83ISNILRR963 pKa = 11.84GTNVAVRR970 pKa = 11.84LDD972 pKa = 3.01IGRR975 pKa = 11.84RR976 pKa = 11.84RR977 pKa = 11.84KK978 pKa = 10.25DD979 pKa = 3.19KK980 pKa = 10.89EE981 pKa = 3.9IVRR984 pKa = 11.84TTLMSHH990 pKa = 6.48RR991 pKa = 11.84SEE993 pKa = 4.27YY994 pKa = 11.11VPMVSANDD1002 pKa = 4.11GSKK1005 pKa = 8.99LTSLIRR1011 pKa = 11.84YY1012 pKa = 7.56DD1013 pKa = 4.01MPTMSGDD1020 pKa = 3.34CGAPLMLSEE1029 pKa = 3.58NRR1031 pKa = 11.84YY1032 pKa = 9.47YY1033 pKa = 10.88GGRR1036 pKa = 11.84CILGLHH1042 pKa = 5.69VAGKK1046 pKa = 5.21TTLMSRR1052 pKa = 11.84EE1053 pKa = 4.52GYY1055 pKa = 9.96SVIASQEE1062 pKa = 3.97CMRR1065 pKa = 11.84EE1066 pKa = 3.13IWLRR1070 pKa = 11.84LGPSEE1075 pKa = 4.14EE1076 pKa = 4.4VEE1078 pKa = 3.88VDD1080 pKa = 3.7TVHH1083 pKa = 6.93SQMRR1087 pKa = 11.84QVTNEE1092 pKa = 3.69EE1093 pKa = 4.47FVTLEE1098 pKa = 3.95AGLVEE1103 pKa = 4.43KK1104 pKa = 10.8GIIGGSMSYY1113 pKa = 10.32LGPLNEE1119 pKa = 4.68PVNLAPKK1126 pKa = 8.76TALIPSPMHH1135 pKa = 6.4ADD1137 pKa = 3.65EE1138 pKa = 5.31PFGPCPVAPAILYY1151 pKa = 8.31PVVKK1155 pKa = 10.27DD1156 pKa = 3.53GLRR1159 pKa = 11.84VYY1161 pKa = 10.78PMARR1165 pKa = 11.84AVEE1168 pKa = 5.14AYY1170 pKa = 10.26QSDD1173 pKa = 4.65VVVKK1177 pKa = 10.34APQVLDD1183 pKa = 3.35IAAEE1187 pKa = 4.3VAFKK1191 pKa = 10.76PLMKK1195 pKa = 9.63VTQDD1199 pKa = 3.31FPRR1202 pKa = 11.84DD1203 pKa = 3.48VLSFEE1208 pKa = 4.4EE1209 pKa = 4.63AVVPPEE1215 pKa = 3.76GWKK1218 pKa = 10.54LKK1220 pKa = 9.63PLNRR1224 pKa = 11.84KK1225 pKa = 9.06SSAGYY1230 pKa = 9.79KK1231 pKa = 8.41YY1232 pKa = 10.37RR1233 pKa = 11.84SYY1235 pKa = 9.99VTAAKK1240 pKa = 9.18PGKK1243 pKa = 9.81VAFLGKK1249 pKa = 10.45EE1250 pKa = 3.72GDD1252 pKa = 3.58VDD1254 pKa = 4.59FSRR1257 pKa = 11.84PEE1259 pKa = 3.63LDD1261 pKa = 3.01IVRR1264 pKa = 11.84KK1265 pKa = 9.8DD1266 pKa = 3.24VTSIISHH1273 pKa = 6.69AKK1275 pKa = 8.92RR1276 pKa = 11.84GVRR1279 pKa = 11.84LPHH1282 pKa = 6.22YY1283 pKa = 9.06CTDD1286 pKa = 3.81FLKK1289 pKa = 11.08DD1290 pKa = 3.39EE1291 pKa = 4.55LRR1293 pKa = 11.84PLEE1296 pKa = 4.18KK1297 pKa = 10.74VEE1299 pKa = 4.0AVKK1302 pKa = 10.05TRR1304 pKa = 11.84MISGTEE1310 pKa = 3.67LDD1312 pKa = 3.44YY1313 pKa = 11.28TIAVRR1318 pKa = 11.84MYY1320 pKa = 9.9FGAFNAAMLATPVVNGMAPGINHH1343 pKa = 4.68YY1344 pKa = 6.97TQWGEE1349 pKa = 3.57LATRR1353 pKa = 11.84LISKK1357 pKa = 10.17GGAVFDD1363 pKa = 4.49GDD1365 pKa = 3.98FSRR1368 pKa = 11.84FDD1370 pKa = 3.43ASEE1373 pKa = 4.1QPWVHH1378 pKa = 5.71MKK1380 pKa = 10.02ILEE1383 pKa = 5.15VINQWYY1389 pKa = 9.62AMKK1392 pKa = 10.7GGTEE1396 pKa = 3.58EE1397 pKa = 4.44DD1398 pKa = 3.47DD1399 pKa = 3.57RR1400 pKa = 11.84VRR1402 pKa = 11.84TILWEE1407 pKa = 4.06DD1408 pKa = 3.91VIHH1411 pKa = 6.23SVHH1414 pKa = 5.73ITGDD1418 pKa = 3.74SSSHH1422 pKa = 5.24GQLVQWHH1429 pKa = 6.6KK1430 pKa = 10.87SLPSGHH1436 pKa = 7.47PLTTVINSMYY1446 pKa = 10.77SLLALTTCYY1455 pKa = 10.32IHH1457 pKa = 6.57LTGDD1461 pKa = 3.42SRR1463 pKa = 11.84DD1464 pKa = 3.28MWEE1467 pKa = 4.15HH1468 pKa = 5.31VFINTFGDD1476 pKa = 3.83DD1477 pKa = 3.29NVAGVDD1483 pKa = 3.51EE1484 pKa = 4.45SVRR1487 pKa = 11.84DD1488 pKa = 3.56VFNQVTVASAMSEE1501 pKa = 4.19LFNLTYY1507 pKa = 9.81TAGAKK1512 pKa = 9.79DD1513 pKa = 3.97GKK1515 pKa = 10.15LVPYY1519 pKa = 9.77TDD1521 pKa = 3.5IYY1523 pKa = 11.18NITFLKK1529 pKa = 10.47RR1530 pKa = 11.84SFLRR1534 pKa = 11.84DD1535 pKa = 3.38EE1536 pKa = 4.26EE1537 pKa = 4.35TSDD1540 pKa = 3.74IIGSAPCMDD1549 pKa = 3.42WVGPLAKK1556 pKa = 10.19EE1557 pKa = 4.1SFLYY1561 pKa = 9.05TPYY1564 pKa = 10.92YY1565 pKa = 9.57YY1566 pKa = 10.41RR1567 pKa = 11.84NKK1569 pKa = 10.08KK1570 pKa = 10.21DD1571 pKa = 3.29PRR1573 pKa = 11.84KK1574 pKa = 10.49DD1575 pKa = 3.0ITDD1578 pKa = 3.43NCDD1581 pKa = 2.96ILLGEE1586 pKa = 4.69LALHH1590 pKa = 6.65PKK1592 pKa = 10.46SMWDD1596 pKa = 3.32EE1597 pKa = 3.96YY1598 pKa = 11.36FPLLKK1603 pKa = 10.03QWCVKK1608 pKa = 10.7NDD1610 pKa = 3.11IEE1612 pKa = 4.53LTFEE1616 pKa = 3.97SRR1618 pKa = 11.84SAARR1622 pKa = 11.84AYY1624 pKa = 8.36ITTRR1628 pKa = 11.84FDD1630 pKa = 2.83VWFF1633 pKa = 4.48
Molecular weight: 182.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2425 |
792 |
1633 |
1212.5 |
133.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.825 ± 0.262 | 1.196 ± 0.094 |
5.402 ± 0.496 | 4.619 ± 0.801 |
4.99 ± 0.923 | 7.381 ± 0.417 |
1.814 ± 0.278 | 4.99 ± 0.033 |
5.361 ± 0.666 | 7.505 ± 0.856 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.175 ± 0.264 | 4.165 ± 0.256 |
4.825 ± 0.624 | 3.052 ± 0.011 |
4.825 ± 0.714 | 7.588 ± 0.886 |
7.216 ± 1.71 | 7.835 ± 0.704 |
1.485 ± 0.176 | 3.753 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
