
Aiptasia sp. sea anemone associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.98
Get precalculated fractions of proteins
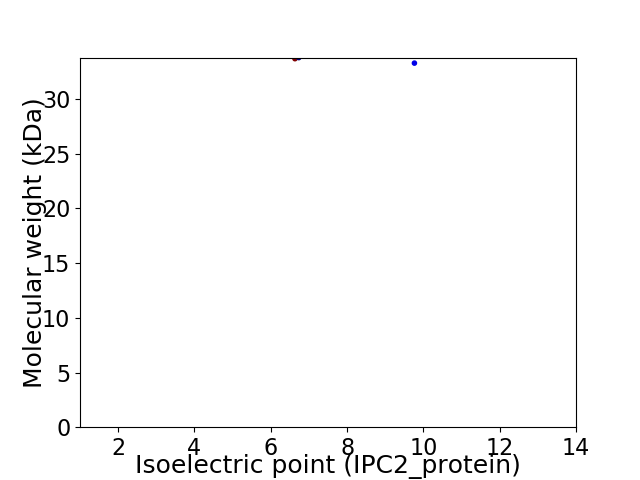
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RL37|A0A0K1RL37_9CIRC Putative replication initiation protein (Fragment) OS=Aiptasia sp. sea anemone associated circular virus OX=1692242 PE=4 SV=1
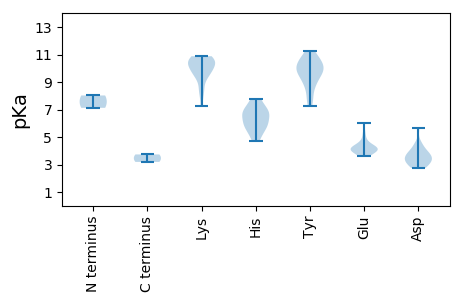
TT1 pKa = 7.12SSSFSSFSHH10 pKa = 7.17DD11 pKa = 2.77AHH13 pKa = 7.62ISIRR17 pKa = 11.84TVVFHH22 pKa = 5.85RR23 pKa = 11.84QWYY26 pKa = 8.52LVVGRR31 pKa = 11.84EE32 pKa = 3.93IGEE35 pKa = 4.05SGTRR39 pKa = 11.84HH40 pKa = 5.1LQGFVIFHH48 pKa = 5.45QVQSRR53 pKa = 11.84AAVSGYY59 pKa = 9.01IARR62 pKa = 11.84AHH64 pKa = 6.58LEE66 pKa = 3.78PARR69 pKa = 11.84ASSVQASDD77 pKa = 3.55YY78 pKa = 10.53CKK80 pKa = 10.48KK81 pKa = 10.85DD82 pKa = 2.95GDD84 pKa = 3.57FDD86 pKa = 4.36EE87 pKa = 6.03YY88 pKa = 11.27GVCPRR93 pKa = 11.84SGQRR97 pKa = 11.84FDD99 pKa = 4.59LQALLKK105 pKa = 10.33WGDD108 pKa = 3.86DD109 pKa = 4.55FIASHH114 pKa = 6.07KK115 pKa = 9.91RR116 pKa = 11.84APTAHH121 pKa = 6.52EE122 pKa = 4.27CAVEE126 pKa = 4.3QPAAYY131 pKa = 9.96LKK133 pKa = 9.77YY134 pKa = 9.81PRR136 pKa = 11.84LISLFQARR144 pKa = 11.84APPPDD149 pKa = 3.86FGRR152 pKa = 11.84EE153 pKa = 4.1GEE155 pKa = 4.07RR156 pKa = 11.84RR157 pKa = 11.84PWQHH161 pKa = 5.71EE162 pKa = 4.17LEE164 pKa = 5.36DD165 pKa = 4.15EE166 pKa = 5.01LEE168 pKa = 4.73GPCEE172 pKa = 3.6NDD174 pKa = 2.79RR175 pKa = 11.84RR176 pKa = 11.84VTFIVDD182 pKa = 3.38QVGGAGKK189 pKa = 8.11TWFQQWFLSKK199 pKa = 10.85NPARR203 pKa = 11.84AQIVSIGKK211 pKa = 9.53RR212 pKa = 11.84DD213 pKa = 3.97DD214 pKa = 3.4VAHH217 pKa = 6.75TIDD220 pKa = 3.33EE221 pKa = 4.43TKK223 pKa = 10.59EE224 pKa = 3.86VFFFAVPRR232 pKa = 11.84GQMEE236 pKa = 4.23FLRR239 pKa = 11.84YY240 pKa = 9.53EE241 pKa = 3.97ILEE244 pKa = 4.07MLKK247 pKa = 10.83DD248 pKa = 3.66RR249 pKa = 11.84MVFSPKK255 pKa = 9.34YY256 pKa = 7.86ASRR259 pKa = 11.84MKK261 pKa = 10.15FLSVVPHH268 pKa = 6.06VIVFSNEE275 pKa = 3.65LPDD278 pKa = 3.85MNKK281 pKa = 9.31MSLDD285 pKa = 3.0RR286 pKa = 11.84YY287 pKa = 9.59YY288 pKa = 10.29IKK290 pKa = 10.53EE291 pKa = 4.13VNN293 pKa = 3.19
TT1 pKa = 7.12SSSFSSFSHH10 pKa = 7.17DD11 pKa = 2.77AHH13 pKa = 7.62ISIRR17 pKa = 11.84TVVFHH22 pKa = 5.85RR23 pKa = 11.84QWYY26 pKa = 8.52LVVGRR31 pKa = 11.84EE32 pKa = 3.93IGEE35 pKa = 4.05SGTRR39 pKa = 11.84HH40 pKa = 5.1LQGFVIFHH48 pKa = 5.45QVQSRR53 pKa = 11.84AAVSGYY59 pKa = 9.01IARR62 pKa = 11.84AHH64 pKa = 6.58LEE66 pKa = 3.78PARR69 pKa = 11.84ASSVQASDD77 pKa = 3.55YY78 pKa = 10.53CKK80 pKa = 10.48KK81 pKa = 10.85DD82 pKa = 2.95GDD84 pKa = 3.57FDD86 pKa = 4.36EE87 pKa = 6.03YY88 pKa = 11.27GVCPRR93 pKa = 11.84SGQRR97 pKa = 11.84FDD99 pKa = 4.59LQALLKK105 pKa = 10.33WGDD108 pKa = 3.86DD109 pKa = 4.55FIASHH114 pKa = 6.07KK115 pKa = 9.91RR116 pKa = 11.84APTAHH121 pKa = 6.52EE122 pKa = 4.27CAVEE126 pKa = 4.3QPAAYY131 pKa = 9.96LKK133 pKa = 9.77YY134 pKa = 9.81PRR136 pKa = 11.84LISLFQARR144 pKa = 11.84APPPDD149 pKa = 3.86FGRR152 pKa = 11.84EE153 pKa = 4.1GEE155 pKa = 4.07RR156 pKa = 11.84RR157 pKa = 11.84PWQHH161 pKa = 5.71EE162 pKa = 4.17LEE164 pKa = 5.36DD165 pKa = 4.15EE166 pKa = 5.01LEE168 pKa = 4.73GPCEE172 pKa = 3.6NDD174 pKa = 2.79RR175 pKa = 11.84RR176 pKa = 11.84VTFIVDD182 pKa = 3.38QVGGAGKK189 pKa = 8.11TWFQQWFLSKK199 pKa = 10.85NPARR203 pKa = 11.84AQIVSIGKK211 pKa = 9.53RR212 pKa = 11.84DD213 pKa = 3.97DD214 pKa = 3.4VAHH217 pKa = 6.75TIDD220 pKa = 3.33EE221 pKa = 4.43TKK223 pKa = 10.59EE224 pKa = 3.86VFFFAVPRR232 pKa = 11.84GQMEE236 pKa = 4.23FLRR239 pKa = 11.84YY240 pKa = 9.53EE241 pKa = 3.97ILEE244 pKa = 4.07MLKK247 pKa = 10.83DD248 pKa = 3.66RR249 pKa = 11.84MVFSPKK255 pKa = 9.34YY256 pKa = 7.86ASRR259 pKa = 11.84MKK261 pKa = 10.15FLSVVPHH268 pKa = 6.06VIVFSNEE275 pKa = 3.65LPDD278 pKa = 3.85MNKK281 pKa = 9.31MSLDD285 pKa = 3.0RR286 pKa = 11.84YY287 pKa = 9.59YY288 pKa = 10.29IKK290 pKa = 10.53EE291 pKa = 4.13VNN293 pKa = 3.19
Molecular weight: 33.7 kDa
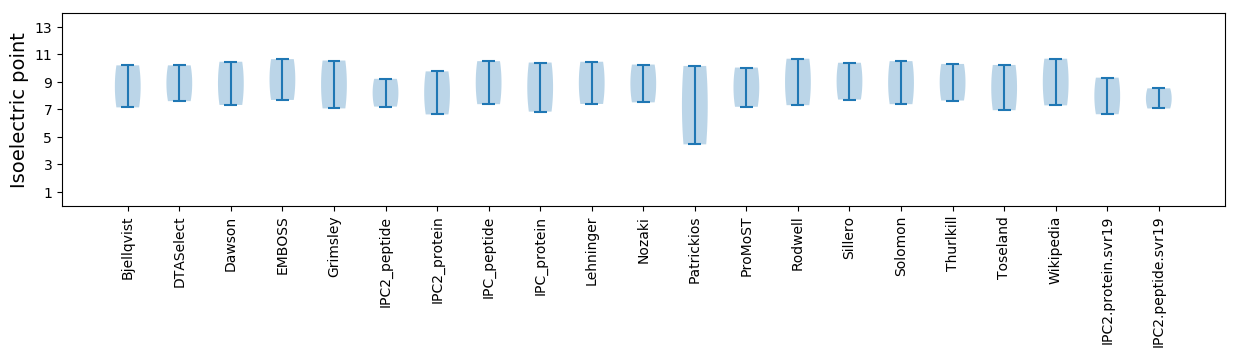
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RL37|A0A0K1RL37_9CIRC Putative replication initiation protein (Fragment) OS=Aiptasia sp. sea anemone associated circular virus OX=1692242 PE=4 SV=1
MM1 pKa = 8.02PYY3 pKa = 10.75GPIVAYY9 pKa = 9.26GAKK12 pKa = 9.81RR13 pKa = 11.84ALIGAAMSTRR23 pKa = 11.84AYY25 pKa = 9.66KK26 pKa = 10.39RR27 pKa = 11.84ARR29 pKa = 11.84TAYY32 pKa = 10.0RR33 pKa = 11.84VGSYY37 pKa = 10.44ARR39 pKa = 11.84AYY41 pKa = 10.15GPTAIRR47 pKa = 11.84AGRR50 pKa = 11.84RR51 pKa = 11.84IWKK54 pKa = 9.61AYY56 pKa = 8.85RR57 pKa = 11.84RR58 pKa = 11.84YY59 pKa = 9.68RR60 pKa = 11.84GKK62 pKa = 10.16NRR64 pKa = 11.84EE65 pKa = 3.89QFSRR69 pKa = 11.84TNIGEE74 pKa = 4.27RR75 pKa = 11.84VGSSSTKK82 pKa = 9.58KK83 pKa = 10.18RR84 pKa = 11.84YY85 pKa = 10.27AFTILNATQGDD96 pKa = 4.13TRR98 pKa = 11.84TLTTHH103 pKa = 7.78LINNIPEE110 pKa = 4.16KK111 pKa = 10.79SALNQNNNRR120 pKa = 11.84RR121 pKa = 11.84RR122 pKa = 11.84QIVNLRR128 pKa = 11.84GVKK131 pKa = 10.1LCLEE135 pKa = 4.61FRR137 pKa = 11.84NQLARR142 pKa = 11.84PLYY145 pKa = 10.82LNVAVLGQKK154 pKa = 7.25TQNQDD159 pKa = 3.11LPPSVLGFFRR169 pKa = 11.84GFDD172 pKa = 3.32QNSRR176 pKa = 11.84SEE178 pKa = 4.41DD179 pKa = 3.25FTTARR184 pKa = 11.84SANEE188 pKa = 3.76LHH190 pKa = 6.8CLPINPDD197 pKa = 2.76KK198 pKa = 10.71FTVLRR203 pKa = 11.84HH204 pKa = 4.72KK205 pKa = 10.44RR206 pKa = 11.84YY207 pKa = 10.0RR208 pKa = 11.84LIPNSDD214 pKa = 3.08AGTVYY219 pKa = 10.63NDD221 pKa = 3.77HH222 pKa = 6.92SGHH225 pKa = 6.88SYY227 pKa = 11.0MNMDD231 pKa = 2.78WWLKK235 pKa = 8.45VKK237 pKa = 10.56RR238 pKa = 11.84QIRR241 pKa = 11.84YY242 pKa = 7.72TDD244 pKa = 3.52GGDD247 pKa = 3.28IDD249 pKa = 5.64SGSLWLCHH257 pKa = 5.86WCDD260 pKa = 3.74EE261 pKa = 4.31IFAGTFAPVQPDD273 pKa = 3.35AYY275 pKa = 9.81AYY277 pKa = 7.28TIRR280 pKa = 11.84SICYY284 pKa = 8.84FKK286 pKa = 10.88EE287 pKa = 4.12PKK289 pKa = 8.57TT290 pKa = 3.73
MM1 pKa = 8.02PYY3 pKa = 10.75GPIVAYY9 pKa = 9.26GAKK12 pKa = 9.81RR13 pKa = 11.84ALIGAAMSTRR23 pKa = 11.84AYY25 pKa = 9.66KK26 pKa = 10.39RR27 pKa = 11.84ARR29 pKa = 11.84TAYY32 pKa = 10.0RR33 pKa = 11.84VGSYY37 pKa = 10.44ARR39 pKa = 11.84AYY41 pKa = 10.15GPTAIRR47 pKa = 11.84AGRR50 pKa = 11.84RR51 pKa = 11.84IWKK54 pKa = 9.61AYY56 pKa = 8.85RR57 pKa = 11.84RR58 pKa = 11.84YY59 pKa = 9.68RR60 pKa = 11.84GKK62 pKa = 10.16NRR64 pKa = 11.84EE65 pKa = 3.89QFSRR69 pKa = 11.84TNIGEE74 pKa = 4.27RR75 pKa = 11.84VGSSSTKK82 pKa = 9.58KK83 pKa = 10.18RR84 pKa = 11.84YY85 pKa = 10.27AFTILNATQGDD96 pKa = 4.13TRR98 pKa = 11.84TLTTHH103 pKa = 7.78LINNIPEE110 pKa = 4.16KK111 pKa = 10.79SALNQNNNRR120 pKa = 11.84RR121 pKa = 11.84RR122 pKa = 11.84QIVNLRR128 pKa = 11.84GVKK131 pKa = 10.1LCLEE135 pKa = 4.61FRR137 pKa = 11.84NQLARR142 pKa = 11.84PLYY145 pKa = 10.82LNVAVLGQKK154 pKa = 7.25TQNQDD159 pKa = 3.11LPPSVLGFFRR169 pKa = 11.84GFDD172 pKa = 3.32QNSRR176 pKa = 11.84SEE178 pKa = 4.41DD179 pKa = 3.25FTTARR184 pKa = 11.84SANEE188 pKa = 3.76LHH190 pKa = 6.8CLPINPDD197 pKa = 2.76KK198 pKa = 10.71FTVLRR203 pKa = 11.84HH204 pKa = 4.72KK205 pKa = 10.44RR206 pKa = 11.84YY207 pKa = 10.0RR208 pKa = 11.84LIPNSDD214 pKa = 3.08AGTVYY219 pKa = 10.63NDD221 pKa = 3.77HH222 pKa = 6.92SGHH225 pKa = 6.88SYY227 pKa = 11.0MNMDD231 pKa = 2.78WWLKK235 pKa = 8.45VKK237 pKa = 10.56RR238 pKa = 11.84QIRR241 pKa = 11.84YY242 pKa = 7.72TDD244 pKa = 3.52GGDD247 pKa = 3.28IDD249 pKa = 5.64SGSLWLCHH257 pKa = 5.86WCDD260 pKa = 3.74EE261 pKa = 4.31IFAGTFAPVQPDD273 pKa = 3.35AYY275 pKa = 9.81AYY277 pKa = 7.28TIRR280 pKa = 11.84SICYY284 pKa = 8.84FKK286 pKa = 10.88EE287 pKa = 4.12PKK289 pKa = 8.57TT290 pKa = 3.73
Molecular weight: 33.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
583 |
290 |
293 |
291.5 |
33.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.233 ± 0.285 | 1.544 ± 0.133 |
5.317 ± 0.614 | 4.803 ± 1.505 |
5.317 ± 1.122 | 6.346 ± 0.405 |
2.916 ± 0.624 | 5.146 ± 0.273 |
4.974 ± 0.146 | 6.69 ± 0.406 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.715 ± 0.247 | 4.117 ± 1.793 |
4.803 ± 0.236 | 4.46 ± 0.491 |
9.605 ± 1.052 | 6.861 ± 0.736 |
4.803 ± 1.542 | 6.003 ± 1.374 |
1.715 ± 0.007 | 4.631 ± 0.906 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
