
Rusa timorensis papillomavirus type 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Treisthetapapillomavirus; Treisthetapapillomavirus 1
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
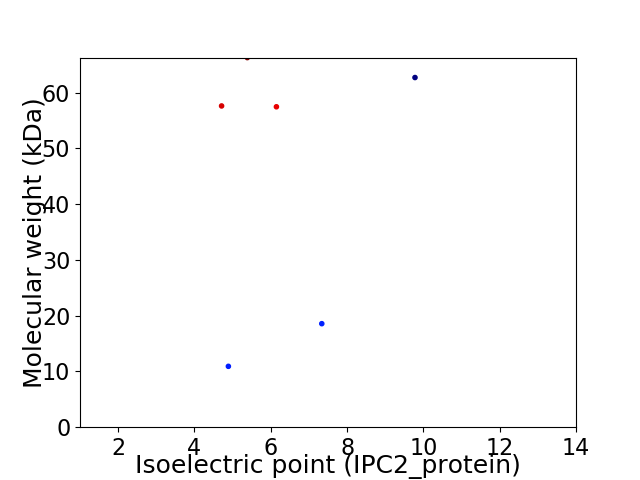
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A109PMF8|A0A109PMF8_9PAPI Major capsid protein L1 OS=Rusa timorensis papillomavirus type 1 OX=1776868 GN=L1 PE=3 SV=1
MM1 pKa = 7.0CAEE4 pKa = 4.76MIGEE8 pKa = 4.3SATIQDD14 pKa = 5.31IILQDD19 pKa = 3.29IDD21 pKa = 5.16SIDD24 pKa = 3.95LACHH28 pKa = 5.32EE29 pKa = 4.52TLSSEE34 pKa = 4.29VEE36 pKa = 3.79QRR38 pKa = 11.84RR39 pKa = 11.84PTVYY43 pKa = 9.56PYY45 pKa = 10.79KK46 pKa = 10.37IVTRR50 pKa = 11.84CGQCEE55 pKa = 3.92TSICLYY61 pKa = 10.65VGSTQIGIVTLQSQLAEE78 pKa = 4.55DD79 pKa = 4.2LCILCGNCGRR89 pKa = 11.84QNFPNGRR96 pKa = 11.84KK97 pKa = 9.56KK98 pKa = 10.74SS99 pKa = 3.63
MM1 pKa = 7.0CAEE4 pKa = 4.76MIGEE8 pKa = 4.3SATIQDD14 pKa = 5.31IILQDD19 pKa = 3.29IDD21 pKa = 5.16SIDD24 pKa = 3.95LACHH28 pKa = 5.32EE29 pKa = 4.52TLSSEE34 pKa = 4.29VEE36 pKa = 3.79QRR38 pKa = 11.84RR39 pKa = 11.84PTVYY43 pKa = 9.56PYY45 pKa = 10.79KK46 pKa = 10.37IVTRR50 pKa = 11.84CGQCEE55 pKa = 3.92TSICLYY61 pKa = 10.65VGSTQIGIVTLQSQLAEE78 pKa = 4.55DD79 pKa = 4.2LCILCGNCGRR89 pKa = 11.84QNFPNGRR96 pKa = 11.84KK97 pKa = 9.56KK98 pKa = 10.74SS99 pKa = 3.63
Molecular weight: 10.92 kDa
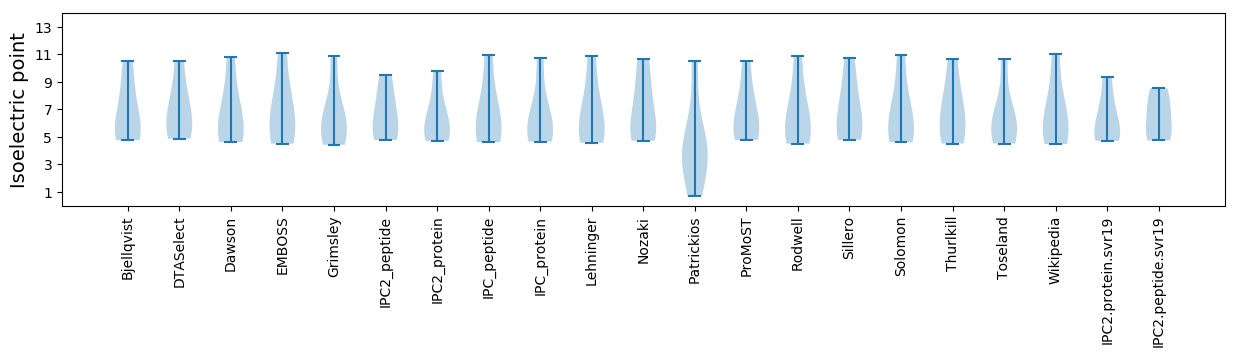
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0X9K182|A0A0X9K182_9PAPI Replication protein E1 OS=Rusa timorensis papillomavirus type 1 OX=1776868 GN=E1 PE=3 SV=1
MM1 pKa = 7.35ATLNDD6 pKa = 4.23RR7 pKa = 11.84FDD9 pKa = 4.89ALQQSQLDD17 pKa = 3.99IIEE20 pKa = 4.91AEE22 pKa = 4.3PGTLEE27 pKa = 4.66DD28 pKa = 5.55QIFYY32 pKa = 10.43WDD34 pKa = 4.17LLRR37 pKa = 11.84QEE39 pKa = 5.61RR40 pKa = 11.84VLQYY44 pKa = 8.74YY45 pKa = 9.91ARR47 pKa = 11.84KK48 pKa = 9.7KK49 pKa = 9.21GYY51 pKa = 9.52KK52 pKa = 9.19RR53 pKa = 11.84LGMLPLPTLQISEE66 pKa = 4.38AAAKK70 pKa = 9.59EE71 pKa = 4.11AIAMTLHH78 pKa = 6.63LSSLKK83 pKa = 10.14EE84 pKa = 3.56SSYY87 pKa = 11.89ANEE90 pKa = 3.76RR91 pKa = 11.84WTLQDD96 pKa = 3.11TSIVTFEE103 pKa = 4.78SPPQFTFKK111 pKa = 10.48KK112 pKa = 10.13GPVAVKK118 pKa = 10.32LSFDD122 pKa = 4.07GNPDD126 pKa = 3.01NAVRR130 pKa = 11.84HH131 pKa = 5.02TLWRR135 pKa = 11.84DD136 pKa = 2.74IYY138 pKa = 11.54YY139 pKa = 10.49LDD141 pKa = 6.46DD142 pKa = 5.24DD143 pKa = 4.42DD144 pKa = 4.39QWRR147 pKa = 11.84KK148 pKa = 7.82TYY150 pKa = 11.18SDD152 pKa = 3.12VDD154 pKa = 3.64SRR156 pKa = 11.84GIYY159 pKa = 8.97YY160 pKa = 10.06WSGDD164 pKa = 3.44SKK166 pKa = 11.34VYY168 pKa = 10.4YY169 pKa = 10.8VEE171 pKa = 4.85FKK173 pKa = 11.03DD174 pKa = 4.63DD175 pKa = 3.51ASLYY179 pKa = 10.44SLTGQWEE186 pKa = 3.99VTYY189 pKa = 10.85NKK191 pKa = 8.97KK192 pKa = 8.31TFSSSIATSSTNEE205 pKa = 3.51EE206 pKa = 4.64DD207 pKa = 3.98GPVPISSDD215 pKa = 3.49EE216 pKa = 4.15EE217 pKa = 4.4GTDD220 pKa = 3.36GHH222 pKa = 5.72GQEE225 pKa = 5.4HH226 pKa = 6.26EE227 pKa = 4.14RR228 pKa = 11.84SPNRR232 pKa = 11.84SDD234 pKa = 3.41TEE236 pKa = 4.06EE237 pKa = 5.25GEE239 pKa = 4.4EE240 pKa = 4.16TGGEE244 pKa = 3.94ASEE247 pKa = 4.45KK248 pKa = 10.61ASISGSVRR256 pKa = 11.84SIRR259 pKa = 11.84TARR262 pKa = 11.84SPKK265 pKa = 9.21AHH267 pKa = 6.6SNPRR271 pKa = 11.84SKK273 pKa = 10.68SRR275 pKa = 11.84SRR277 pKa = 11.84SRR279 pKa = 11.84SRR281 pKa = 11.84SRR283 pKa = 11.84SSTRR287 pKa = 11.84GWTRR291 pKa = 11.84ARR293 pKa = 11.84QTSSSGSRR301 pKa = 11.84SRR303 pKa = 11.84SRR305 pKa = 11.84SGSRR309 pKa = 11.84TRR311 pKa = 11.84ARR313 pKa = 11.84SWRR316 pKa = 11.84RR317 pKa = 11.84SPGHH321 pKa = 6.57RR322 pKa = 11.84GRR324 pKa = 11.84GRR326 pKa = 11.84GRR328 pKa = 11.84PPGSGRR334 pKa = 11.84RR335 pKa = 11.84QGGAGSRR342 pKa = 11.84PGAGSGAGGRR352 pKa = 11.84PGLRR356 pKa = 11.84SRR358 pKa = 11.84SRR360 pKa = 11.84DD361 pKa = 3.11TRR363 pKa = 11.84AGSGLPNRR371 pKa = 11.84PSASPRR377 pKa = 11.84AEE379 pKa = 3.53PVRR382 pKa = 11.84VLEE385 pKa = 4.16GSIPRR390 pKa = 11.84PSSYY394 pKa = 11.14GLRR397 pKa = 11.84GSGGEE402 pKa = 4.3TEE404 pKa = 5.14APTDD408 pKa = 3.57TSSLVSPPHH417 pKa = 6.06LSPWFRR423 pKa = 11.84DD424 pKa = 3.72PPRR427 pKa = 11.84APGRR431 pKa = 11.84PPSGRR436 pKa = 11.84HH437 pKa = 5.01RR438 pKa = 11.84RR439 pKa = 11.84PAPTPEE445 pKa = 3.85RR446 pKa = 11.84RR447 pKa = 11.84TTSPLARR454 pKa = 11.84RR455 pKa = 11.84KK456 pKa = 8.82RR457 pKa = 11.84TGSRR461 pKa = 11.84STEE464 pKa = 4.01RR465 pKa = 11.84LHH467 pKa = 6.37STPTSAKK474 pKa = 9.25KK475 pKa = 10.17DD476 pKa = 3.44PPVIIVRR483 pKa = 11.84GLPNQLKK490 pKa = 6.72TWRR493 pKa = 11.84YY494 pKa = 8.18RR495 pKa = 11.84LHH497 pKa = 6.3NRR499 pKa = 11.84RR500 pKa = 11.84KK501 pKa = 9.76KK502 pKa = 10.96LPFLYY507 pKa = 10.7LSTTFSWVTRR517 pKa = 11.84KK518 pKa = 10.63GGDD521 pKa = 3.22RR522 pKa = 11.84LSSTRR527 pKa = 11.84MLVAFASYY535 pKa = 10.95SDD537 pKa = 3.06RR538 pKa = 11.84SYY540 pKa = 11.45FLRR543 pKa = 11.84HH544 pKa = 4.92VVFPPGVTFSLGYY557 pKa = 10.24FYY559 pKa = 11.49GLL561 pKa = 3.62
MM1 pKa = 7.35ATLNDD6 pKa = 4.23RR7 pKa = 11.84FDD9 pKa = 4.89ALQQSQLDD17 pKa = 3.99IIEE20 pKa = 4.91AEE22 pKa = 4.3PGTLEE27 pKa = 4.66DD28 pKa = 5.55QIFYY32 pKa = 10.43WDD34 pKa = 4.17LLRR37 pKa = 11.84QEE39 pKa = 5.61RR40 pKa = 11.84VLQYY44 pKa = 8.74YY45 pKa = 9.91ARR47 pKa = 11.84KK48 pKa = 9.7KK49 pKa = 9.21GYY51 pKa = 9.52KK52 pKa = 9.19RR53 pKa = 11.84LGMLPLPTLQISEE66 pKa = 4.38AAAKK70 pKa = 9.59EE71 pKa = 4.11AIAMTLHH78 pKa = 6.63LSSLKK83 pKa = 10.14EE84 pKa = 3.56SSYY87 pKa = 11.89ANEE90 pKa = 3.76RR91 pKa = 11.84WTLQDD96 pKa = 3.11TSIVTFEE103 pKa = 4.78SPPQFTFKK111 pKa = 10.48KK112 pKa = 10.13GPVAVKK118 pKa = 10.32LSFDD122 pKa = 4.07GNPDD126 pKa = 3.01NAVRR130 pKa = 11.84HH131 pKa = 5.02TLWRR135 pKa = 11.84DD136 pKa = 2.74IYY138 pKa = 11.54YY139 pKa = 10.49LDD141 pKa = 6.46DD142 pKa = 5.24DD143 pKa = 4.42DD144 pKa = 4.39QWRR147 pKa = 11.84KK148 pKa = 7.82TYY150 pKa = 11.18SDD152 pKa = 3.12VDD154 pKa = 3.64SRR156 pKa = 11.84GIYY159 pKa = 8.97YY160 pKa = 10.06WSGDD164 pKa = 3.44SKK166 pKa = 11.34VYY168 pKa = 10.4YY169 pKa = 10.8VEE171 pKa = 4.85FKK173 pKa = 11.03DD174 pKa = 4.63DD175 pKa = 3.51ASLYY179 pKa = 10.44SLTGQWEE186 pKa = 3.99VTYY189 pKa = 10.85NKK191 pKa = 8.97KK192 pKa = 8.31TFSSSIATSSTNEE205 pKa = 3.51EE206 pKa = 4.64DD207 pKa = 3.98GPVPISSDD215 pKa = 3.49EE216 pKa = 4.15EE217 pKa = 4.4GTDD220 pKa = 3.36GHH222 pKa = 5.72GQEE225 pKa = 5.4HH226 pKa = 6.26EE227 pKa = 4.14RR228 pKa = 11.84SPNRR232 pKa = 11.84SDD234 pKa = 3.41TEE236 pKa = 4.06EE237 pKa = 5.25GEE239 pKa = 4.4EE240 pKa = 4.16TGGEE244 pKa = 3.94ASEE247 pKa = 4.45KK248 pKa = 10.61ASISGSVRR256 pKa = 11.84SIRR259 pKa = 11.84TARR262 pKa = 11.84SPKK265 pKa = 9.21AHH267 pKa = 6.6SNPRR271 pKa = 11.84SKK273 pKa = 10.68SRR275 pKa = 11.84SRR277 pKa = 11.84SRR279 pKa = 11.84SRR281 pKa = 11.84SRR283 pKa = 11.84SSTRR287 pKa = 11.84GWTRR291 pKa = 11.84ARR293 pKa = 11.84QTSSSGSRR301 pKa = 11.84SRR303 pKa = 11.84SRR305 pKa = 11.84SGSRR309 pKa = 11.84TRR311 pKa = 11.84ARR313 pKa = 11.84SWRR316 pKa = 11.84RR317 pKa = 11.84SPGHH321 pKa = 6.57RR322 pKa = 11.84GRR324 pKa = 11.84GRR326 pKa = 11.84GRR328 pKa = 11.84PPGSGRR334 pKa = 11.84RR335 pKa = 11.84QGGAGSRR342 pKa = 11.84PGAGSGAGGRR352 pKa = 11.84PGLRR356 pKa = 11.84SRR358 pKa = 11.84SRR360 pKa = 11.84DD361 pKa = 3.11TRR363 pKa = 11.84AGSGLPNRR371 pKa = 11.84PSASPRR377 pKa = 11.84AEE379 pKa = 3.53PVRR382 pKa = 11.84VLEE385 pKa = 4.16GSIPRR390 pKa = 11.84PSSYY394 pKa = 11.14GLRR397 pKa = 11.84GSGGEE402 pKa = 4.3TEE404 pKa = 5.14APTDD408 pKa = 3.57TSSLVSPPHH417 pKa = 6.06LSPWFRR423 pKa = 11.84DD424 pKa = 3.72PPRR427 pKa = 11.84APGRR431 pKa = 11.84PPSGRR436 pKa = 11.84HH437 pKa = 5.01RR438 pKa = 11.84RR439 pKa = 11.84PAPTPEE445 pKa = 3.85RR446 pKa = 11.84RR447 pKa = 11.84TTSPLARR454 pKa = 11.84RR455 pKa = 11.84KK456 pKa = 8.82RR457 pKa = 11.84TGSRR461 pKa = 11.84STEE464 pKa = 4.01RR465 pKa = 11.84LHH467 pKa = 6.37STPTSAKK474 pKa = 9.25KK475 pKa = 10.17DD476 pKa = 3.44PPVIIVRR483 pKa = 11.84GLPNQLKK490 pKa = 6.72TWRR493 pKa = 11.84YY494 pKa = 8.18RR495 pKa = 11.84LHH497 pKa = 6.3NRR499 pKa = 11.84RR500 pKa = 11.84KK501 pKa = 9.76KK502 pKa = 10.96LPFLYY507 pKa = 10.7LSTTFSWVTRR517 pKa = 11.84KK518 pKa = 10.63GGDD521 pKa = 3.22RR522 pKa = 11.84LSSTRR527 pKa = 11.84MLVAFASYY535 pKa = 10.95SDD537 pKa = 3.06RR538 pKa = 11.84SYY540 pKa = 11.45FLRR543 pKa = 11.84HH544 pKa = 4.92VVFPPGVTFSLGYY557 pKa = 10.24FYY559 pKa = 11.49GLL561 pKa = 3.62
Molecular weight: 62.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2437 |
99 |
585 |
406.2 |
45.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.032 ± 0.783 | 2.134 ± 0.933 |
6.483 ± 0.425 | 5.868 ± 0.381 |
4.637 ± 0.603 | 7.591 ± 0.783 |
2.093 ± 0.255 | 4.432 ± 0.83 |
4.76 ± 0.974 | 7.92 ± 0.597 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.395 ± 0.21 | 3.57 ± 0.427 |
6.319 ± 0.637 | 3.816 ± 0.391 |
7.427 ± 1.33 | 8.33 ± 1.421 |
6.155 ± 0.558 | 5.909 ± 0.595 |
1.477 ± 0.314 | 3.652 ± 0.34 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
