
Tadarida brasiliensis circovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; Bat associated circovirus 4
Average proteome isoelectric point is 8.67
Get precalculated fractions of proteins
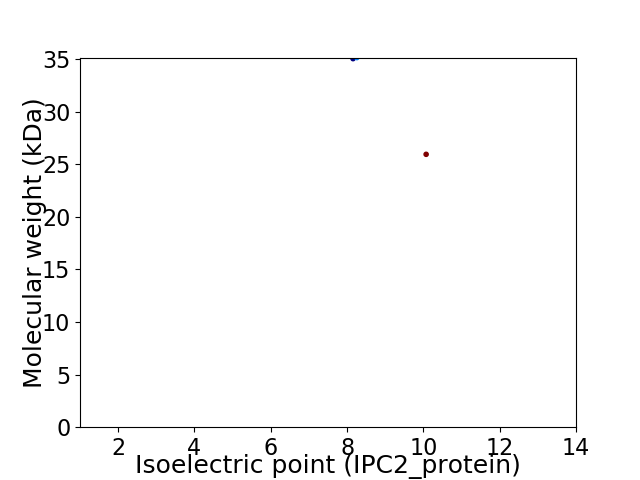
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
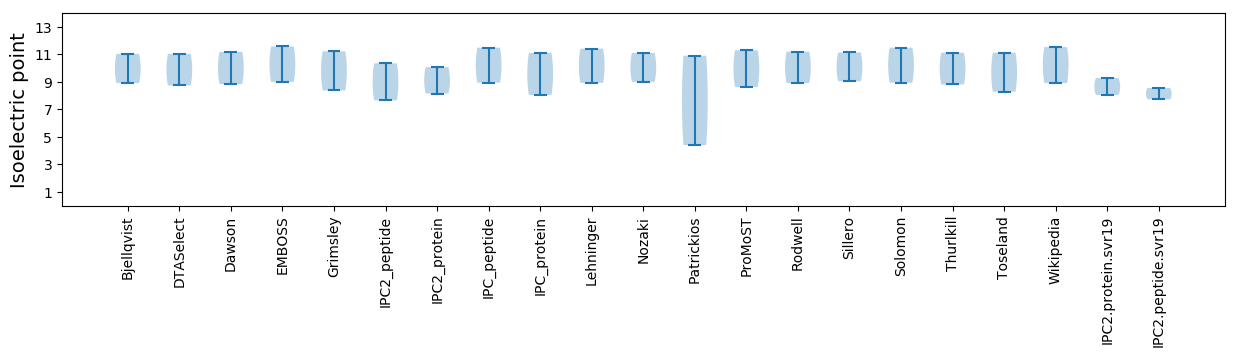
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0N9P0D8|A0A0N9P0D8_9CIRC Capsid protein OS=Tadarida brasiliensis circovirus 1 OX=1732201 GN=cap PE=3 SV=1
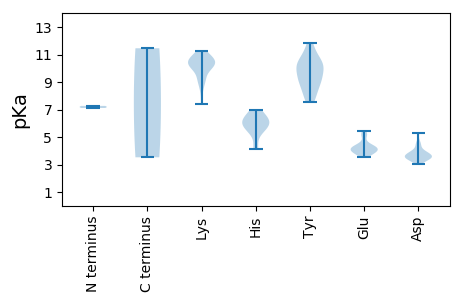
MM1 pKa = 7.11NVKK4 pKa = 10.23RR5 pKa = 11.84KK6 pKa = 9.86YY7 pKa = 9.09NAKK10 pKa = 10.4KK11 pKa = 10.43GDD13 pKa = 3.72EE14 pKa = 4.37TPGRR18 pKa = 11.84RR19 pKa = 11.84WCFTINNYY27 pKa = 8.5GAPDD31 pKa = 3.99LEE33 pKa = 4.73CVNEE37 pKa = 3.95AFRR40 pKa = 11.84CEE42 pKa = 4.03DD43 pKa = 3.05VVYY46 pKa = 10.3AICGKK51 pKa = 10.31EE52 pKa = 3.9KK53 pKa = 10.22GAKK56 pKa = 7.39GTPHH60 pKa = 6.27LQGFIHH66 pKa = 6.44FTGNWRR72 pKa = 11.84FNRR75 pKa = 11.84VRR77 pKa = 11.84NLLGGRR83 pKa = 11.84AHH85 pKa = 6.94IEE87 pKa = 3.84KK88 pKa = 10.73ARR90 pKa = 11.84GNDD93 pKa = 3.78DD94 pKa = 3.3QNKK97 pKa = 9.03AYY99 pKa = 10.16CSKK102 pKa = 10.83EE103 pKa = 3.66EE104 pKa = 4.14TYY106 pKa = 11.25LEE108 pKa = 4.24VGTPQFQGKK117 pKa = 9.93RR118 pKa = 11.84NDD120 pKa = 3.5LGRR123 pKa = 11.84VASALEE129 pKa = 4.16SGATLSEE136 pKa = 4.32VARR139 pKa = 11.84ASPEE143 pKa = 3.6VFIRR147 pKa = 11.84YY148 pKa = 8.71GRR150 pKa = 11.84GLRR153 pKa = 11.84DD154 pKa = 3.38YY155 pKa = 9.8MNVRR159 pKa = 11.84GLVKK163 pKa = 10.32PRR165 pKa = 11.84DD166 pKa = 3.78FKK168 pKa = 11.2TEE170 pKa = 4.0VIVLVGEE177 pKa = 4.32PGSGKK182 pKa = 10.21SKK184 pKa = 10.51YY185 pKa = 10.56ANEE188 pKa = 4.4LPGSKK193 pKa = 9.24YY194 pKa = 9.34WKK196 pKa = 9.87PRR198 pKa = 11.84GQWWDD203 pKa = 3.52GYY205 pKa = 9.64NGEE208 pKa = 5.22DD209 pKa = 4.59IVVLDD214 pKa = 5.32DD215 pKa = 3.98FYY217 pKa = 11.83GWVPYY222 pKa = 10.67DD223 pKa = 3.39EE224 pKa = 5.15LLRR227 pKa = 11.84IGDD230 pKa = 4.37RR231 pKa = 11.84YY232 pKa = 8.95PLKK235 pKa = 10.68VQVKK239 pKa = 8.31GAFVEE244 pKa = 5.43FTSKK248 pKa = 10.21MLVITSNKK256 pKa = 9.43RR257 pKa = 11.84PEE259 pKa = 3.55EE260 pKa = 4.22WYY262 pKa = 10.23DD263 pKa = 3.27KK264 pKa = 11.23EE265 pKa = 5.15KK266 pKa = 10.78IADD269 pKa = 3.62QSAMWRR275 pKa = 11.84RR276 pKa = 11.84FDD278 pKa = 3.87KK279 pKa = 10.3MYY281 pKa = 9.39YY282 pKa = 9.26CEE284 pKa = 4.17RR285 pKa = 11.84GEE287 pKa = 3.93PVKK290 pKa = 10.55AYY292 pKa = 8.08PHH294 pKa = 5.71EE295 pKa = 4.24WKK297 pKa = 10.61EE298 pKa = 3.93FEE300 pKa = 3.95TGYY303 pKa = 11.46
MM1 pKa = 7.11NVKK4 pKa = 10.23RR5 pKa = 11.84KK6 pKa = 9.86YY7 pKa = 9.09NAKK10 pKa = 10.4KK11 pKa = 10.43GDD13 pKa = 3.72EE14 pKa = 4.37TPGRR18 pKa = 11.84RR19 pKa = 11.84WCFTINNYY27 pKa = 8.5GAPDD31 pKa = 3.99LEE33 pKa = 4.73CVNEE37 pKa = 3.95AFRR40 pKa = 11.84CEE42 pKa = 4.03DD43 pKa = 3.05VVYY46 pKa = 10.3AICGKK51 pKa = 10.31EE52 pKa = 3.9KK53 pKa = 10.22GAKK56 pKa = 7.39GTPHH60 pKa = 6.27LQGFIHH66 pKa = 6.44FTGNWRR72 pKa = 11.84FNRR75 pKa = 11.84VRR77 pKa = 11.84NLLGGRR83 pKa = 11.84AHH85 pKa = 6.94IEE87 pKa = 3.84KK88 pKa = 10.73ARR90 pKa = 11.84GNDD93 pKa = 3.78DD94 pKa = 3.3QNKK97 pKa = 9.03AYY99 pKa = 10.16CSKK102 pKa = 10.83EE103 pKa = 3.66EE104 pKa = 4.14TYY106 pKa = 11.25LEE108 pKa = 4.24VGTPQFQGKK117 pKa = 9.93RR118 pKa = 11.84NDD120 pKa = 3.5LGRR123 pKa = 11.84VASALEE129 pKa = 4.16SGATLSEE136 pKa = 4.32VARR139 pKa = 11.84ASPEE143 pKa = 3.6VFIRR147 pKa = 11.84YY148 pKa = 8.71GRR150 pKa = 11.84GLRR153 pKa = 11.84DD154 pKa = 3.38YY155 pKa = 9.8MNVRR159 pKa = 11.84GLVKK163 pKa = 10.32PRR165 pKa = 11.84DD166 pKa = 3.78FKK168 pKa = 11.2TEE170 pKa = 4.0VIVLVGEE177 pKa = 4.32PGSGKK182 pKa = 10.21SKK184 pKa = 10.51YY185 pKa = 10.56ANEE188 pKa = 4.4LPGSKK193 pKa = 9.24YY194 pKa = 9.34WKK196 pKa = 9.87PRR198 pKa = 11.84GQWWDD203 pKa = 3.52GYY205 pKa = 9.64NGEE208 pKa = 5.22DD209 pKa = 4.59IVVLDD214 pKa = 5.32DD215 pKa = 3.98FYY217 pKa = 11.83GWVPYY222 pKa = 10.67DD223 pKa = 3.39EE224 pKa = 5.15LLRR227 pKa = 11.84IGDD230 pKa = 4.37RR231 pKa = 11.84YY232 pKa = 8.95PLKK235 pKa = 10.68VQVKK239 pKa = 8.31GAFVEE244 pKa = 5.43FTSKK248 pKa = 10.21MLVITSNKK256 pKa = 9.43RR257 pKa = 11.84PEE259 pKa = 3.55EE260 pKa = 4.22WYY262 pKa = 10.23DD263 pKa = 3.27KK264 pKa = 11.23EE265 pKa = 5.15KK266 pKa = 10.78IADD269 pKa = 3.62QSAMWRR275 pKa = 11.84RR276 pKa = 11.84FDD278 pKa = 3.87KK279 pKa = 10.3MYY281 pKa = 9.39YY282 pKa = 9.26CEE284 pKa = 4.17RR285 pKa = 11.84GEE287 pKa = 3.93PVKK290 pKa = 10.55AYY292 pKa = 8.08PHH294 pKa = 5.71EE295 pKa = 4.24WKK297 pKa = 10.61EE298 pKa = 3.93FEE300 pKa = 3.95TGYY303 pKa = 11.46
Molecular weight: 35.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0N9P0D8|A0A0N9P0D8_9CIRC Capsid protein OS=Tadarida brasiliensis circovirus 1 OX=1732201 GN=cap PE=3 SV=1
MM1 pKa = 7.27PRR3 pKa = 11.84SRR5 pKa = 11.84RR6 pKa = 11.84HH7 pKa = 4.13RR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84NQWFKK16 pKa = 10.05RR17 pKa = 11.84WRR19 pKa = 11.84RR20 pKa = 11.84SRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84GHH26 pKa = 5.42TFGTRR31 pKa = 11.84RR32 pKa = 11.84WRR34 pKa = 11.84QKK36 pKa = 10.18NGIYY40 pKa = 10.3NFKK43 pKa = 10.63FKK45 pKa = 11.02ANSLQTVNKK54 pKa = 9.06TSNQGYY60 pKa = 7.73FTFSLAAACPRR71 pKa = 11.84AFSTYY76 pKa = 9.1FDD78 pKa = 3.56MYY80 pKa = 10.15RR81 pKa = 11.84IRR83 pKa = 11.84KK84 pKa = 8.71VRR86 pKa = 11.84VQWLPMSGINEE97 pKa = 3.84PRR99 pKa = 11.84VWGATIIDD107 pKa = 3.72LTGRR111 pKa = 11.84DD112 pKa = 3.44TTLPSTGNTDD122 pKa = 3.67FTIDD126 pKa = 3.59DD127 pKa = 3.55VTRR130 pKa = 11.84RR131 pKa = 11.84IWNPTRR137 pKa = 11.84LHH139 pKa = 5.86SRR141 pKa = 11.84YY142 pKa = 7.56FTPKK146 pKa = 9.85PEE148 pKa = 3.95IAIKK152 pKa = 9.27TVSSEE157 pKa = 4.31AIQPNNPRR165 pKa = 11.84NQLWLDD171 pKa = 3.45AKK173 pKa = 11.12YY174 pKa = 10.59PDD176 pKa = 4.51AKK178 pKa = 10.7HH179 pKa = 6.27HH180 pKa = 5.94GCAFFFGQTDD190 pKa = 3.91LGDD193 pKa = 3.53EE194 pKa = 4.33SYY196 pKa = 11.31KK197 pKa = 10.73FQFLVTYY204 pKa = 8.22YY205 pKa = 10.02IQFRR209 pKa = 11.84QFAGSLAPSS218 pKa = 3.52
MM1 pKa = 7.27PRR3 pKa = 11.84SRR5 pKa = 11.84RR6 pKa = 11.84HH7 pKa = 4.13RR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84NQWFKK16 pKa = 10.05RR17 pKa = 11.84WRR19 pKa = 11.84RR20 pKa = 11.84SRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84GHH26 pKa = 5.42TFGTRR31 pKa = 11.84RR32 pKa = 11.84WRR34 pKa = 11.84QKK36 pKa = 10.18NGIYY40 pKa = 10.3NFKK43 pKa = 10.63FKK45 pKa = 11.02ANSLQTVNKK54 pKa = 9.06TSNQGYY60 pKa = 7.73FTFSLAAACPRR71 pKa = 11.84AFSTYY76 pKa = 9.1FDD78 pKa = 3.56MYY80 pKa = 10.15RR81 pKa = 11.84IRR83 pKa = 11.84KK84 pKa = 8.71VRR86 pKa = 11.84VQWLPMSGINEE97 pKa = 3.84PRR99 pKa = 11.84VWGATIIDD107 pKa = 3.72LTGRR111 pKa = 11.84DD112 pKa = 3.44TTLPSTGNTDD122 pKa = 3.67FTIDD126 pKa = 3.59DD127 pKa = 3.55VTRR130 pKa = 11.84RR131 pKa = 11.84IWNPTRR137 pKa = 11.84LHH139 pKa = 5.86SRR141 pKa = 11.84YY142 pKa = 7.56FTPKK146 pKa = 9.85PEE148 pKa = 3.95IAIKK152 pKa = 9.27TVSSEE157 pKa = 4.31AIQPNNPRR165 pKa = 11.84NQLWLDD171 pKa = 3.45AKK173 pKa = 11.12YY174 pKa = 10.59PDD176 pKa = 4.51AKK178 pKa = 10.7HH179 pKa = 6.27HH180 pKa = 5.94GCAFFFGQTDD190 pKa = 3.91LGDD193 pKa = 3.53EE194 pKa = 4.33SYY196 pKa = 11.31KK197 pKa = 10.73FQFLVTYY204 pKa = 8.22YY205 pKa = 10.02IQFRR209 pKa = 11.84QFAGSLAPSS218 pKa = 3.52
Molecular weight: 25.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
521 |
218 |
303 |
260.5 |
30.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.95 ± 0.008 | 1.536 ± 0.377 |
5.182 ± 0.363 | 5.758 ± 2.393 |
5.758 ± 1.245 | 8.061 ± 1.56 |
1.727 ± 0.345 | 4.031 ± 0.619 |
7.102 ± 1.254 | 5.374 ± 0.2 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.536 ± 0.097 | 5.182 ± 0.197 |
4.99 ± 0.314 | 3.455 ± 0.971 |
9.789 ± 1.584 | 4.798 ± 0.99 |
5.758 ± 1.804 | 5.566 ± 1.437 |
3.263 ± 0.248 | 5.182 ± 0.643 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
