
Amphibola crenata associated bacilladnavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Baphyvirales; Bacilladnaviridae; Protobacilladnavirus; Snail associated protobacilladnavirus 1
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
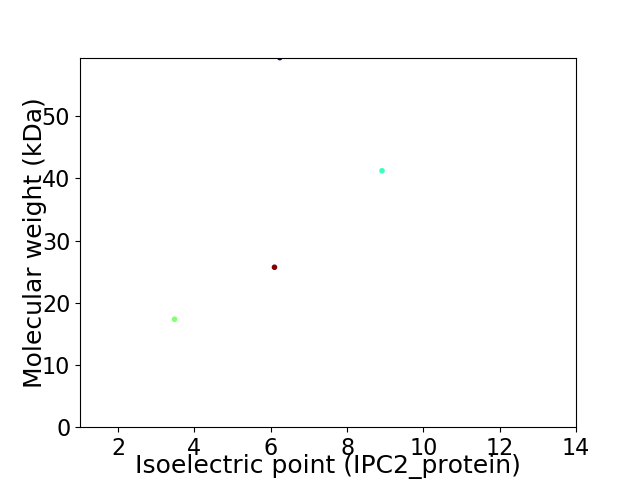
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1P8YT79|A0A1P8YT79_9VIRU p2 OS=Amphibola crenata associated bacilladnavirus 1 OX=1941435 PE=4 SV=1
MM1 pKa = 7.71AIINTHH7 pKa = 5.69MPFSDD12 pKa = 3.54NGEE15 pKa = 4.4LIQVDD20 pKa = 3.83MQKK23 pKa = 11.1ALSCLMFFNTLGIYY37 pKa = 10.5NDD39 pKa = 3.09ATYY42 pKa = 11.09YY43 pKa = 10.68GIKK46 pKa = 10.12LWMDD50 pKa = 3.44AYY52 pKa = 8.78MTIGNAPMPSGVLFKK67 pKa = 11.06SPLGGYY73 pKa = 9.69VGFTQLMAEE82 pKa = 5.26LEE84 pKa = 4.41QEE86 pKa = 3.9WAARR90 pKa = 11.84EE91 pKa = 4.12AFHH94 pKa = 6.46LTIDD98 pKa = 3.9EE99 pKa = 4.66EE100 pKa = 4.38EE101 pKa = 4.36VDD103 pKa = 4.27DD104 pKa = 6.29DD105 pKa = 4.45EE106 pKa = 4.92STVTGPVTQPEE117 pKa = 4.49YY118 pKa = 10.97EE119 pKa = 4.5PYY121 pKa = 11.09ALDD124 pKa = 5.37DD125 pKa = 5.19DD126 pKa = 5.31DD127 pKa = 7.36SIDD130 pKa = 3.46AMLNIPVNIDD140 pKa = 3.23FNLGFFDD147 pKa = 5.25EE148 pKa = 4.52MDD150 pKa = 3.77EE151 pKa = 4.32EE152 pKa = 4.75DD153 pKa = 3.61VV154 pKa = 3.85
MM1 pKa = 7.71AIINTHH7 pKa = 5.69MPFSDD12 pKa = 3.54NGEE15 pKa = 4.4LIQVDD20 pKa = 3.83MQKK23 pKa = 11.1ALSCLMFFNTLGIYY37 pKa = 10.5NDD39 pKa = 3.09ATYY42 pKa = 11.09YY43 pKa = 10.68GIKK46 pKa = 10.12LWMDD50 pKa = 3.44AYY52 pKa = 8.78MTIGNAPMPSGVLFKK67 pKa = 11.06SPLGGYY73 pKa = 9.69VGFTQLMAEE82 pKa = 5.26LEE84 pKa = 4.41QEE86 pKa = 3.9WAARR90 pKa = 11.84EE91 pKa = 4.12AFHH94 pKa = 6.46LTIDD98 pKa = 3.9EE99 pKa = 4.66EE100 pKa = 4.38EE101 pKa = 4.36VDD103 pKa = 4.27DD104 pKa = 6.29DD105 pKa = 4.45EE106 pKa = 4.92STVTGPVTQPEE117 pKa = 4.49YY118 pKa = 10.97EE119 pKa = 4.5PYY121 pKa = 11.09ALDD124 pKa = 5.37DD125 pKa = 5.19DD126 pKa = 5.31DD127 pKa = 7.36SIDD130 pKa = 3.46AMLNIPVNIDD140 pKa = 3.23FNLGFFDD147 pKa = 5.25EE148 pKa = 4.52MDD150 pKa = 3.77EE151 pKa = 4.32EE152 pKa = 4.75DD153 pKa = 3.61VV154 pKa = 3.85
Molecular weight: 17.36 kDa
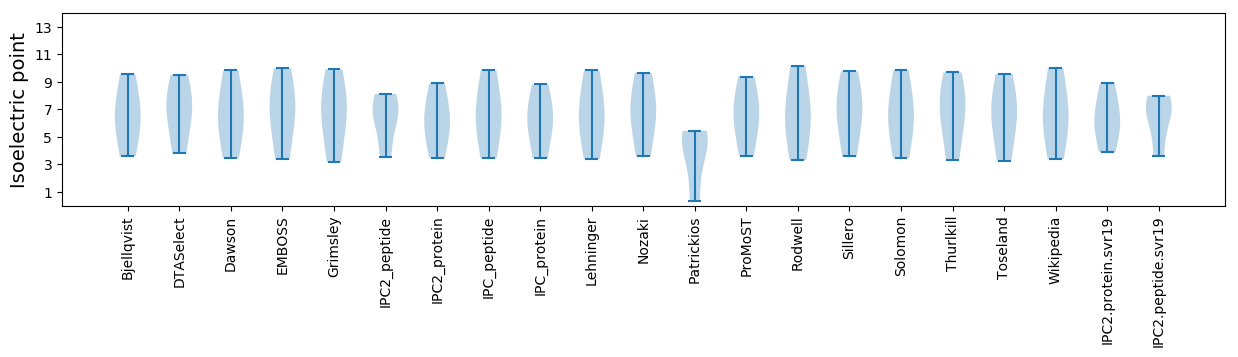
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8YT82|A0A1P8YT82_9VIRU p1 OS=Amphibola crenata associated bacilladnavirus 1 OX=1941435 PE=4 SV=1
MM1 pKa = 7.61PKK3 pKa = 9.88KK4 pKa = 9.88SSRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84TTATTKK16 pKa = 10.45KK17 pKa = 10.09KK18 pKa = 10.62SSSKK22 pKa = 10.14KK23 pKa = 9.03AAKK26 pKa = 9.89KK27 pKa = 10.67GPTTKK32 pKa = 10.52KK33 pKa = 9.64VVKK36 pKa = 10.13KK37 pKa = 11.03VMGASAIALKK47 pKa = 10.43PLVSPFDD54 pKa = 4.04RR55 pKa = 11.84SNMQPKK61 pKa = 10.01IPDD64 pKa = 3.59GKK66 pKa = 10.06VGEE69 pKa = 4.41SVGLKK74 pKa = 8.92FTMTKK79 pKa = 10.01EE80 pKa = 4.12YY81 pKa = 11.02SNKK84 pKa = 9.18TLTTMHH90 pKa = 6.8AMLFPGQSGGLFVSGCQNVNGINRR114 pKa = 11.84EE115 pKa = 3.95VQAYY119 pKa = 9.63ANAGGFSYY127 pKa = 10.14TYY129 pKa = 11.44DD130 pKa = 3.59NTTKK134 pKa = 10.15MALLTANEE142 pKa = 5.22KK143 pKa = 8.12YY144 pKa = 10.35HH145 pKa = 5.35SWRR148 pKa = 11.84IVSQAAKK155 pKa = 10.61FEE157 pKa = 4.38LLNPAEE163 pKa = 4.99EE164 pKa = 4.94DD165 pKa = 3.82DD166 pKa = 4.36GWWEE170 pKa = 3.92AVRR173 pKa = 11.84ITDD176 pKa = 3.57MLSLFDD182 pKa = 3.65YY183 pKa = 10.72HH184 pKa = 8.42VEE186 pKa = 3.82QHH188 pKa = 5.71SNKK191 pKa = 9.19TGRR194 pKa = 11.84LQGVFVPFLGLLNLEE209 pKa = 4.55DD210 pKa = 3.75RR211 pKa = 11.84ALTAEE216 pKa = 4.02RR217 pKa = 11.84TYY219 pKa = 11.35EE220 pKa = 3.95SGRR223 pKa = 11.84LKK225 pKa = 10.54DD226 pKa = 2.96IHH228 pKa = 7.59KK229 pKa = 10.52RR230 pKa = 11.84EE231 pKa = 3.73FHH233 pKa = 5.66LRR235 pKa = 11.84QVKK238 pKa = 10.85DD239 pKa = 3.55EE240 pKa = 4.14VDD242 pKa = 4.08FTQLCSPIALEE253 pKa = 4.67GVQEE257 pKa = 4.36TGAGTSGSPFVLANGAGANAGEE279 pKa = 4.35SFGMQNYY286 pKa = 9.46NAMRR290 pKa = 11.84NYY292 pKa = 9.84IDD294 pKa = 4.92FGYY297 pKa = 10.76DD298 pKa = 3.22CIYY301 pKa = 10.8FRR303 pKa = 11.84FHH305 pKa = 6.77CRR307 pKa = 11.84DD308 pKa = 3.22PASATRR314 pKa = 11.84LLTHH318 pKa = 6.1VVSNQEE324 pKa = 3.41IQYY327 pKa = 9.84SSEE330 pKa = 3.95SGEE333 pKa = 4.83SRR335 pKa = 11.84FQTANTRR342 pKa = 11.84HH343 pKa = 5.51AQVDD347 pKa = 3.81TVMANRR353 pKa = 11.84SNGGATTSVPSTSRR367 pKa = 11.84HH368 pKa = 4.66NPHH371 pKa = 6.91RR372 pKa = 4.38
MM1 pKa = 7.61PKK3 pKa = 9.88KK4 pKa = 9.88SSRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84TTATTKK16 pKa = 10.45KK17 pKa = 10.09KK18 pKa = 10.62SSSKK22 pKa = 10.14KK23 pKa = 9.03AAKK26 pKa = 9.89KK27 pKa = 10.67GPTTKK32 pKa = 10.52KK33 pKa = 9.64VVKK36 pKa = 10.13KK37 pKa = 11.03VMGASAIALKK47 pKa = 10.43PLVSPFDD54 pKa = 4.04RR55 pKa = 11.84SNMQPKK61 pKa = 10.01IPDD64 pKa = 3.59GKK66 pKa = 10.06VGEE69 pKa = 4.41SVGLKK74 pKa = 8.92FTMTKK79 pKa = 10.01EE80 pKa = 4.12YY81 pKa = 11.02SNKK84 pKa = 9.18TLTTMHH90 pKa = 6.8AMLFPGQSGGLFVSGCQNVNGINRR114 pKa = 11.84EE115 pKa = 3.95VQAYY119 pKa = 9.63ANAGGFSYY127 pKa = 10.14TYY129 pKa = 11.44DD130 pKa = 3.59NTTKK134 pKa = 10.15MALLTANEE142 pKa = 5.22KK143 pKa = 8.12YY144 pKa = 10.35HH145 pKa = 5.35SWRR148 pKa = 11.84IVSQAAKK155 pKa = 10.61FEE157 pKa = 4.38LLNPAEE163 pKa = 4.99EE164 pKa = 4.94DD165 pKa = 3.82DD166 pKa = 4.36GWWEE170 pKa = 3.92AVRR173 pKa = 11.84ITDD176 pKa = 3.57MLSLFDD182 pKa = 3.65YY183 pKa = 10.72HH184 pKa = 8.42VEE186 pKa = 3.82QHH188 pKa = 5.71SNKK191 pKa = 9.19TGRR194 pKa = 11.84LQGVFVPFLGLLNLEE209 pKa = 4.55DD210 pKa = 3.75RR211 pKa = 11.84ALTAEE216 pKa = 4.02RR217 pKa = 11.84TYY219 pKa = 11.35EE220 pKa = 3.95SGRR223 pKa = 11.84LKK225 pKa = 10.54DD226 pKa = 2.96IHH228 pKa = 7.59KK229 pKa = 10.52RR230 pKa = 11.84EE231 pKa = 3.73FHH233 pKa = 5.66LRR235 pKa = 11.84QVKK238 pKa = 10.85DD239 pKa = 3.55EE240 pKa = 4.14VDD242 pKa = 4.08FTQLCSPIALEE253 pKa = 4.67GVQEE257 pKa = 4.36TGAGTSGSPFVLANGAGANAGEE279 pKa = 4.35SFGMQNYY286 pKa = 9.46NAMRR290 pKa = 11.84NYY292 pKa = 9.84IDD294 pKa = 4.92FGYY297 pKa = 10.76DD298 pKa = 3.22CIYY301 pKa = 10.8FRR303 pKa = 11.84FHH305 pKa = 6.77CRR307 pKa = 11.84DD308 pKa = 3.22PASATRR314 pKa = 11.84LLTHH318 pKa = 6.1VVSNQEE324 pKa = 3.41IQYY327 pKa = 9.84SSEE330 pKa = 3.95SGEE333 pKa = 4.83SRR335 pKa = 11.84FQTANTRR342 pKa = 11.84HH343 pKa = 5.51AQVDD347 pKa = 3.81TVMANRR353 pKa = 11.84SNGGATTSVPSTSRR367 pKa = 11.84HH368 pKa = 4.66NPHH371 pKa = 6.91RR372 pKa = 4.38
Molecular weight: 41.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1260 |
154 |
513 |
315.0 |
35.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.952 ± 0.985 | 1.111 ± 0.344 |
7.381 ± 1.687 | 6.905 ± 0.784 |
3.81 ± 0.64 | 6.111 ± 0.659 |
3.254 ± 0.841 | 4.524 ± 0.68 |
7.46 ± 1.393 | 5.714 ± 0.754 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.095 ± 0.614 | 4.841 ± 0.661 |
5.397 ± 0.629 | 3.81 ± 0.131 |
5.397 ± 0.803 | 6.587 ± 0.797 |
6.825 ± 0.734 | 6.27 ± 0.584 |
2.063 ± 0.64 | 3.492 ± 0.212 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
