
Odonata-associated circular virus-9
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.83
Get precalculated fractions of proteins
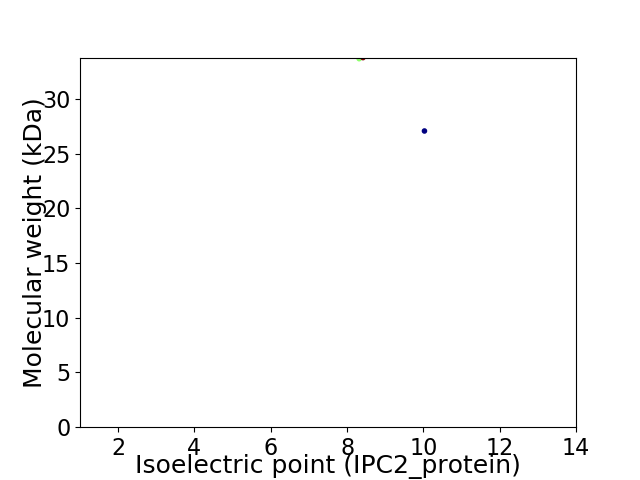
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UI03|A0A0B4UI03_9VIRU Putative capsid protein OS=Odonata-associated circular virus-9 OX=1592129 PE=4 SV=1
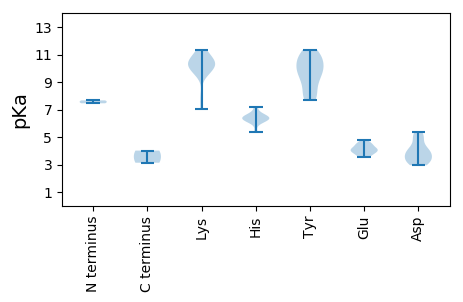
MM1 pKa = 7.45IFKK4 pKa = 10.58KK5 pKa = 10.38KK6 pKa = 10.05IYY8 pKa = 9.89FSNKK12 pKa = 7.93IKK14 pKa = 10.17MSSRR18 pKa = 11.84NYY20 pKa = 10.38VLTIYY25 pKa = 10.64DD26 pKa = 3.84FNLLPKK32 pKa = 10.1LQDD35 pKa = 3.15SAIRR39 pKa = 11.84YY40 pKa = 7.69SAYY43 pKa = 9.47TDD45 pKa = 4.3EE46 pKa = 4.82ICPTTGRR53 pKa = 11.84PHH55 pKa = 5.34KK56 pKa = 9.79QAYY59 pKa = 7.93IVLYY63 pKa = 10.0KK64 pKa = 10.06KK65 pKa = 10.18QRR67 pKa = 11.84PSFVRR72 pKa = 11.84KK73 pKa = 10.04LFPNTYY79 pKa = 7.99NHH81 pKa = 6.38IMNGTLEE88 pKa = 4.02QNDD91 pKa = 3.49IYY93 pKa = 10.67ISKK96 pKa = 10.76KK97 pKa = 7.94NTQQIHH103 pKa = 6.18EE104 pKa = 4.53YY105 pKa = 9.69GDD107 pKa = 3.34KK108 pKa = 11.1PMTQEE113 pKa = 3.82EE114 pKa = 4.46KK115 pKa = 10.91GEE117 pKa = 3.88EE118 pKa = 4.09GAAYY122 pKa = 9.0WRR124 pKa = 11.84EE125 pKa = 3.83QLEE128 pKa = 4.25LAKK131 pKa = 10.04KK132 pKa = 10.41DD133 pKa = 4.09PSLCDD138 pKa = 3.35PKK140 pKa = 11.02LQIIHH145 pKa = 6.39HH146 pKa = 6.67RR147 pKa = 11.84NLEE150 pKa = 4.48AIHH153 pKa = 6.31EE154 pKa = 4.2KK155 pKa = 10.49AKK157 pKa = 10.36RR158 pKa = 11.84VKK160 pKa = 10.78LSDD163 pKa = 4.73LDD165 pKa = 5.2KK166 pKa = 11.35LDD168 pKa = 3.24NWWYY172 pKa = 10.0IGCTGTGKK180 pKa = 10.36SRR182 pKa = 11.84TARR185 pKa = 11.84QLYY188 pKa = 7.94PDD190 pKa = 5.05AYY192 pKa = 9.77IKK194 pKa = 10.4QCNKK198 pKa = 7.01WWNDD202 pKa = 3.04YY203 pKa = 11.11DD204 pKa = 4.3GEE206 pKa = 4.35EE207 pKa = 4.07TVIIDD212 pKa = 5.38DD213 pKa = 4.99LNPDD217 pKa = 3.93HH218 pKa = 7.18KK219 pKa = 11.29FMANFIDD226 pKa = 3.86QWCDD230 pKa = 2.95HH231 pKa = 6.49YY232 pKa = 11.34PFPAEE237 pKa = 3.92FKK239 pKa = 10.57GGQKK243 pKa = 9.58TIRR246 pKa = 11.84PKK248 pKa = 10.71RR249 pKa = 11.84FIITTRR255 pKa = 11.84YY256 pKa = 7.83TIEE259 pKa = 4.63EE260 pKa = 3.89ILGEE264 pKa = 4.2NNFSQIEE271 pKa = 4.05SWKK274 pKa = 10.3RR275 pKa = 11.84RR276 pKa = 11.84FQIKK280 pKa = 9.79EE281 pKa = 3.73FF282 pKa = 4.02
MM1 pKa = 7.45IFKK4 pKa = 10.58KK5 pKa = 10.38KK6 pKa = 10.05IYY8 pKa = 9.89FSNKK12 pKa = 7.93IKK14 pKa = 10.17MSSRR18 pKa = 11.84NYY20 pKa = 10.38VLTIYY25 pKa = 10.64DD26 pKa = 3.84FNLLPKK32 pKa = 10.1LQDD35 pKa = 3.15SAIRR39 pKa = 11.84YY40 pKa = 7.69SAYY43 pKa = 9.47TDD45 pKa = 4.3EE46 pKa = 4.82ICPTTGRR53 pKa = 11.84PHH55 pKa = 5.34KK56 pKa = 9.79QAYY59 pKa = 7.93IVLYY63 pKa = 10.0KK64 pKa = 10.06KK65 pKa = 10.18QRR67 pKa = 11.84PSFVRR72 pKa = 11.84KK73 pKa = 10.04LFPNTYY79 pKa = 7.99NHH81 pKa = 6.38IMNGTLEE88 pKa = 4.02QNDD91 pKa = 3.49IYY93 pKa = 10.67ISKK96 pKa = 10.76KK97 pKa = 7.94NTQQIHH103 pKa = 6.18EE104 pKa = 4.53YY105 pKa = 9.69GDD107 pKa = 3.34KK108 pKa = 11.1PMTQEE113 pKa = 3.82EE114 pKa = 4.46KK115 pKa = 10.91GEE117 pKa = 3.88EE118 pKa = 4.09GAAYY122 pKa = 9.0WRR124 pKa = 11.84EE125 pKa = 3.83QLEE128 pKa = 4.25LAKK131 pKa = 10.04KK132 pKa = 10.41DD133 pKa = 4.09PSLCDD138 pKa = 3.35PKK140 pKa = 11.02LQIIHH145 pKa = 6.39HH146 pKa = 6.67RR147 pKa = 11.84NLEE150 pKa = 4.48AIHH153 pKa = 6.31EE154 pKa = 4.2KK155 pKa = 10.49AKK157 pKa = 10.36RR158 pKa = 11.84VKK160 pKa = 10.78LSDD163 pKa = 4.73LDD165 pKa = 5.2KK166 pKa = 11.35LDD168 pKa = 3.24NWWYY172 pKa = 10.0IGCTGTGKK180 pKa = 10.36SRR182 pKa = 11.84TARR185 pKa = 11.84QLYY188 pKa = 7.94PDD190 pKa = 5.05AYY192 pKa = 9.77IKK194 pKa = 10.4QCNKK198 pKa = 7.01WWNDD202 pKa = 3.04YY203 pKa = 11.11DD204 pKa = 4.3GEE206 pKa = 4.35EE207 pKa = 4.07TVIIDD212 pKa = 5.38DD213 pKa = 4.99LNPDD217 pKa = 3.93HH218 pKa = 7.18KK219 pKa = 11.29FMANFIDD226 pKa = 3.86QWCDD230 pKa = 2.95HH231 pKa = 6.49YY232 pKa = 11.34PFPAEE237 pKa = 3.92FKK239 pKa = 10.57GGQKK243 pKa = 9.58TIRR246 pKa = 11.84PKK248 pKa = 10.71RR249 pKa = 11.84FIITTRR255 pKa = 11.84YY256 pKa = 7.83TIEE259 pKa = 4.63EE260 pKa = 3.89ILGEE264 pKa = 4.2NNFSQIEE271 pKa = 4.05SWKK274 pKa = 10.3RR275 pKa = 11.84RR276 pKa = 11.84FQIKK280 pKa = 9.79EE281 pKa = 3.73FF282 pKa = 4.02
Molecular weight: 33.68 kDa
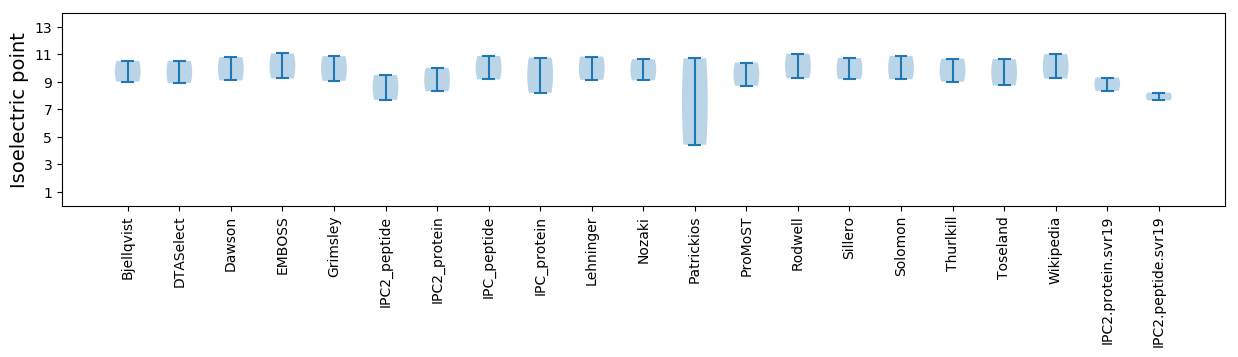
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UI03|A0A0B4UI03_9VIRU Putative capsid protein OS=Odonata-associated circular virus-9 OX=1592129 PE=4 SV=1
MM1 pKa = 7.66ARR3 pKa = 11.84RR4 pKa = 11.84KK5 pKa = 9.53YY6 pKa = 10.48FRR8 pKa = 11.84RR9 pKa = 11.84NPYY12 pKa = 8.93RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FKK17 pKa = 10.32RR18 pKa = 11.84RR19 pKa = 11.84NYY21 pKa = 9.25RR22 pKa = 11.84FSSRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84SGFRR32 pKa = 11.84RR33 pKa = 11.84WRR35 pKa = 11.84VARR38 pKa = 11.84QKK40 pKa = 11.33NLVFSNSQLVTLRR53 pKa = 11.84YY54 pKa = 9.57CKK56 pKa = 10.47NIQLNPANGATISVATFRR74 pKa = 11.84ANDD77 pKa = 3.13IYY79 pKa = 11.12FCDD82 pKa = 4.0GLAGSEE88 pKa = 4.06PKK90 pKa = 9.91GTTEE94 pKa = 3.55WEE96 pKa = 4.16GFYY99 pKa = 11.04NHH101 pKa = 6.45YY102 pKa = 10.7LVIGSKK108 pKa = 7.14MTAMFNSGSSTTSNGAGTVGVVLTDD133 pKa = 3.63TSNVAYY139 pKa = 9.92INNTDD144 pKa = 3.25VMTEE148 pKa = 3.56PRR150 pKa = 11.84ARR152 pKa = 11.84WKK154 pKa = 10.59IIGNQTGTGTKK165 pKa = 9.53TITKK169 pKa = 9.56GYY171 pKa = 8.75SPKK174 pKa = 10.66KK175 pKa = 10.06FFTIHH180 pKa = 6.5NLLDD184 pKa = 3.79NQEE187 pKa = 4.52HH188 pKa = 6.13YY189 pKa = 10.24GAPVSVSPPEE199 pKa = 3.54IAFFQVFFAPHH210 pKa = 6.22AASAGTGGPWVTVIIDD226 pKa = 3.73YY227 pKa = 10.92KK228 pKa = 11.31VMYY231 pKa = 8.56TGPKK235 pKa = 9.62RR236 pKa = 11.84IAPSS240 pKa = 3.13
MM1 pKa = 7.66ARR3 pKa = 11.84RR4 pKa = 11.84KK5 pKa = 9.53YY6 pKa = 10.48FRR8 pKa = 11.84RR9 pKa = 11.84NPYY12 pKa = 8.93RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FKK17 pKa = 10.32RR18 pKa = 11.84RR19 pKa = 11.84NYY21 pKa = 9.25RR22 pKa = 11.84FSSRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84SGFRR32 pKa = 11.84RR33 pKa = 11.84WRR35 pKa = 11.84VARR38 pKa = 11.84QKK40 pKa = 11.33NLVFSNSQLVTLRR53 pKa = 11.84YY54 pKa = 9.57CKK56 pKa = 10.47NIQLNPANGATISVATFRR74 pKa = 11.84ANDD77 pKa = 3.13IYY79 pKa = 11.12FCDD82 pKa = 4.0GLAGSEE88 pKa = 4.06PKK90 pKa = 9.91GTTEE94 pKa = 3.55WEE96 pKa = 4.16GFYY99 pKa = 11.04NHH101 pKa = 6.45YY102 pKa = 10.7LVIGSKK108 pKa = 7.14MTAMFNSGSSTTSNGAGTVGVVLTDD133 pKa = 3.63TSNVAYY139 pKa = 9.92INNTDD144 pKa = 3.25VMTEE148 pKa = 3.56PRR150 pKa = 11.84ARR152 pKa = 11.84WKK154 pKa = 10.59IIGNQTGTGTKK165 pKa = 9.53TITKK169 pKa = 9.56GYY171 pKa = 8.75SPKK174 pKa = 10.66KK175 pKa = 10.06FFTIHH180 pKa = 6.5NLLDD184 pKa = 3.79NQEE187 pKa = 4.52HH188 pKa = 6.13YY189 pKa = 10.24GAPVSVSPPEE199 pKa = 3.54IAFFQVFFAPHH210 pKa = 6.22AASAGTGGPWVTVIIDD226 pKa = 3.73YY227 pKa = 10.92KK228 pKa = 11.31VMYY231 pKa = 8.56TGPKK235 pKa = 9.62RR236 pKa = 11.84IAPSS240 pKa = 3.13
Molecular weight: 27.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
522 |
240 |
282 |
261.0 |
30.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.747 ± 1.189 | 1.341 ± 0.344 |
4.598 ± 1.423 | 4.789 ± 1.553 |
5.364 ± 0.601 | 6.13 ± 1.494 |
2.299 ± 0.429 | 7.471 ± 1.394 |
8.238 ± 1.913 | 5.172 ± 0.965 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.916 ± 0.114 | 6.322 ± 0.516 |
4.789 ± 0.143 | 4.023 ± 1.033 |
7.088 ± 1.41 | 5.747 ± 1.189 |
7.28 ± 1.28 | 4.023 ± 1.793 |
2.107 ± 0.299 | 5.556 ± 0.377 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
