
Lysinibacillus telephonicus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Lysinibacillus
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
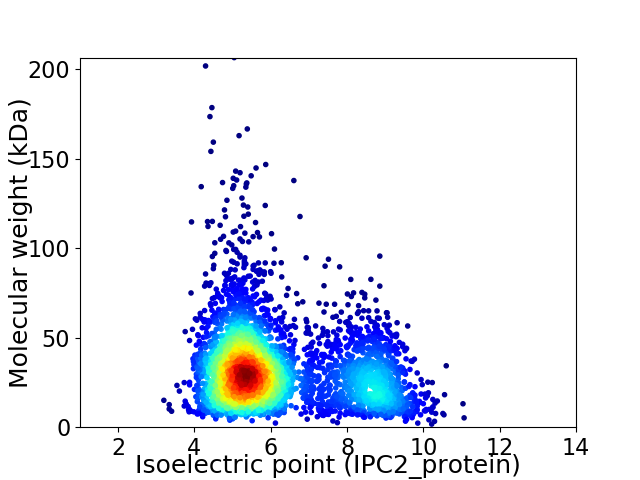
Virtual 2D-PAGE plot for 3959 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S0J0Z7|A0A3S0J0Z7_9BACI Uncharacterized protein OS=Lysinibacillus telephonicus OX=1714840 GN=EKG35_13430 PE=4 SV=1
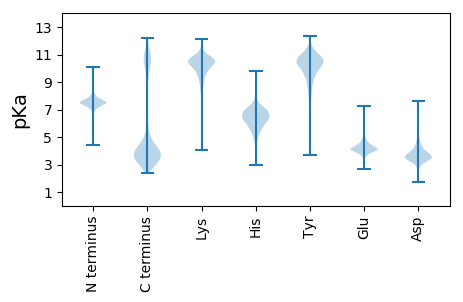
MM1 pKa = 7.5KK2 pKa = 10.64KK3 pKa = 10.43LGTFLLSFSLLIALFPQITMAAQDD27 pKa = 3.73TNFEE31 pKa = 3.99QDD33 pKa = 3.91LINYY37 pKa = 8.58LEE39 pKa = 4.08QVSNEE44 pKa = 3.82RR45 pKa = 11.84GFEE48 pKa = 4.05VTKK51 pKa = 10.89EE52 pKa = 3.99DD53 pKa = 3.29IALSLSYY60 pKa = 11.38YY61 pKa = 10.33EE62 pKa = 6.01DD63 pKa = 3.18SLEE66 pKa = 4.17NYY68 pKa = 7.66NTVEE72 pKa = 4.22EE73 pKa = 4.4LSQFLGEE80 pKa = 4.65IIKK83 pKa = 10.49ADD85 pKa = 3.73YY86 pKa = 11.33SNVDD90 pKa = 3.67SIYY93 pKa = 10.15EE94 pKa = 3.96QYY96 pKa = 11.29EE97 pKa = 4.01LTEE100 pKa = 4.08EE101 pKa = 4.63SLVQLLKK108 pKa = 11.18EE109 pKa = 4.16NGEE112 pKa = 4.05QLSDD116 pKa = 4.17YY117 pKa = 11.03IFLDD121 pKa = 4.08DD122 pKa = 5.9LDD124 pKa = 4.17TAVYY128 pKa = 9.29FYY130 pKa = 11.01TEE132 pKa = 4.58DD133 pKa = 3.45GTFEE137 pKa = 4.43RR138 pKa = 11.84DD139 pKa = 3.19PNFEE143 pKa = 4.14QNLIDD148 pKa = 3.96YY149 pKa = 9.52LADD152 pKa = 3.35VSDD155 pKa = 3.47QRR157 pKa = 11.84GYY159 pKa = 11.34NVTRR163 pKa = 11.84DD164 pKa = 3.53NVVNSLAIYY173 pKa = 9.39EE174 pKa = 4.47SNLEE178 pKa = 4.02DD179 pKa = 4.08FEE181 pKa = 5.54SVEE184 pKa = 3.99QLSDD188 pKa = 3.43FLGEE192 pKa = 4.39VIQADD197 pKa = 4.56YY198 pKa = 11.76SNLDD202 pKa = 3.54YY203 pKa = 10.84FVEE206 pKa = 4.5NYY208 pKa = 10.44GLSQQEE214 pKa = 4.31LFNLLEE220 pKa = 4.72SNDD223 pKa = 3.88EE224 pKa = 4.42NINDD228 pKa = 4.07YY229 pKa = 10.91IYY231 pKa = 10.46IDD233 pKa = 4.78DD234 pKa = 5.47LDD236 pKa = 3.89NTVWKK241 pKa = 10.85YY242 pKa = 10.94VGNDD246 pKa = 3.25LSVMFEE252 pKa = 4.28YY253 pKa = 9.69LTEE256 pKa = 4.71DD257 pKa = 4.16LLPLFQEE264 pKa = 4.05IDD266 pKa = 3.53LTDD269 pKa = 3.69EE270 pKa = 3.7EE271 pKa = 4.66LKK273 pKa = 10.85RR274 pKa = 11.84FEE276 pKa = 3.99NHH278 pKa = 7.06LISLEE283 pKa = 4.04DD284 pKa = 3.4HH285 pKa = 6.77FSNPEE290 pKa = 3.81TLEE293 pKa = 4.06LLEE296 pKa = 3.96QLNYY300 pKa = 10.65EE301 pKa = 4.77LIALEE306 pKa = 4.49SDD308 pKa = 3.76MNSQPTVEE316 pKa = 4.01QMEE319 pKa = 4.29MSLSILEE326 pKa = 4.64KK327 pKa = 10.34IFSILQLKK335 pKa = 10.24ASYY338 pKa = 11.07SLVNDD343 pKa = 5.27AIEE346 pKa = 4.33TPISLMDD353 pKa = 3.88LMKK356 pKa = 10.24MEE358 pKa = 4.29VLEE361 pKa = 4.4GTDD364 pKa = 3.6LKK366 pKa = 11.33VAFYY370 pKa = 10.78SSEE373 pKa = 3.95GQFLGDD379 pKa = 4.55LIITEE384 pKa = 4.19EE385 pKa = 4.57FINSGLVTDD394 pKa = 5.03LGEE397 pKa = 4.28QIVDD401 pKa = 3.86SAVEE405 pKa = 4.01QPPVVQPKK413 pKa = 9.87KK414 pKa = 10.52QINTTQTLAEE424 pKa = 4.58EE425 pKa = 4.33YY426 pKa = 8.8HH427 pKa = 5.2TVKK430 pKa = 10.59GGKK433 pKa = 9.0LPKK436 pKa = 9.21TASDD440 pKa = 4.27YY441 pKa = 11.22IPNALFGLFIVMLGIFMYY459 pKa = 10.58GKK461 pKa = 10.2VRR463 pKa = 11.84TSS465 pKa = 2.77
MM1 pKa = 7.5KK2 pKa = 10.64KK3 pKa = 10.43LGTFLLSFSLLIALFPQITMAAQDD27 pKa = 3.73TNFEE31 pKa = 3.99QDD33 pKa = 3.91LINYY37 pKa = 8.58LEE39 pKa = 4.08QVSNEE44 pKa = 3.82RR45 pKa = 11.84GFEE48 pKa = 4.05VTKK51 pKa = 10.89EE52 pKa = 3.99DD53 pKa = 3.29IALSLSYY60 pKa = 11.38YY61 pKa = 10.33EE62 pKa = 6.01DD63 pKa = 3.18SLEE66 pKa = 4.17NYY68 pKa = 7.66NTVEE72 pKa = 4.22EE73 pKa = 4.4LSQFLGEE80 pKa = 4.65IIKK83 pKa = 10.49ADD85 pKa = 3.73YY86 pKa = 11.33SNVDD90 pKa = 3.67SIYY93 pKa = 10.15EE94 pKa = 3.96QYY96 pKa = 11.29EE97 pKa = 4.01LTEE100 pKa = 4.08EE101 pKa = 4.63SLVQLLKK108 pKa = 11.18EE109 pKa = 4.16NGEE112 pKa = 4.05QLSDD116 pKa = 4.17YY117 pKa = 11.03IFLDD121 pKa = 4.08DD122 pKa = 5.9LDD124 pKa = 4.17TAVYY128 pKa = 9.29FYY130 pKa = 11.01TEE132 pKa = 4.58DD133 pKa = 3.45GTFEE137 pKa = 4.43RR138 pKa = 11.84DD139 pKa = 3.19PNFEE143 pKa = 4.14QNLIDD148 pKa = 3.96YY149 pKa = 9.52LADD152 pKa = 3.35VSDD155 pKa = 3.47QRR157 pKa = 11.84GYY159 pKa = 11.34NVTRR163 pKa = 11.84DD164 pKa = 3.53NVVNSLAIYY173 pKa = 9.39EE174 pKa = 4.47SNLEE178 pKa = 4.02DD179 pKa = 4.08FEE181 pKa = 5.54SVEE184 pKa = 3.99QLSDD188 pKa = 3.43FLGEE192 pKa = 4.39VIQADD197 pKa = 4.56YY198 pKa = 11.76SNLDD202 pKa = 3.54YY203 pKa = 10.84FVEE206 pKa = 4.5NYY208 pKa = 10.44GLSQQEE214 pKa = 4.31LFNLLEE220 pKa = 4.72SNDD223 pKa = 3.88EE224 pKa = 4.42NINDD228 pKa = 4.07YY229 pKa = 10.91IYY231 pKa = 10.46IDD233 pKa = 4.78DD234 pKa = 5.47LDD236 pKa = 3.89NTVWKK241 pKa = 10.85YY242 pKa = 10.94VGNDD246 pKa = 3.25LSVMFEE252 pKa = 4.28YY253 pKa = 9.69LTEE256 pKa = 4.71DD257 pKa = 4.16LLPLFQEE264 pKa = 4.05IDD266 pKa = 3.53LTDD269 pKa = 3.69EE270 pKa = 3.7EE271 pKa = 4.66LKK273 pKa = 10.85RR274 pKa = 11.84FEE276 pKa = 3.99NHH278 pKa = 7.06LISLEE283 pKa = 4.04DD284 pKa = 3.4HH285 pKa = 6.77FSNPEE290 pKa = 3.81TLEE293 pKa = 4.06LLEE296 pKa = 3.96QLNYY300 pKa = 10.65EE301 pKa = 4.77LIALEE306 pKa = 4.49SDD308 pKa = 3.76MNSQPTVEE316 pKa = 4.01QMEE319 pKa = 4.29MSLSILEE326 pKa = 4.64KK327 pKa = 10.34IFSILQLKK335 pKa = 10.24ASYY338 pKa = 11.07SLVNDD343 pKa = 5.27AIEE346 pKa = 4.33TPISLMDD353 pKa = 3.88LMKK356 pKa = 10.24MEE358 pKa = 4.29VLEE361 pKa = 4.4GTDD364 pKa = 3.6LKK366 pKa = 11.33VAFYY370 pKa = 10.78SSEE373 pKa = 3.95GQFLGDD379 pKa = 4.55LIITEE384 pKa = 4.19EE385 pKa = 4.57FINSGLVTDD394 pKa = 5.03LGEE397 pKa = 4.28QIVDD401 pKa = 3.86SAVEE405 pKa = 4.01QPPVVQPKK413 pKa = 9.87KK414 pKa = 10.52QINTTQTLAEE424 pKa = 4.58EE425 pKa = 4.33YY426 pKa = 8.8HH427 pKa = 5.2TVKK430 pKa = 10.59GGKK433 pKa = 9.0LPKK436 pKa = 9.21TASDD440 pKa = 4.27YY441 pKa = 11.22IPNALFGLFIVMLGIFMYY459 pKa = 10.58GKK461 pKa = 10.2VRR463 pKa = 11.84TSS465 pKa = 2.77
Molecular weight: 53.37 kDa
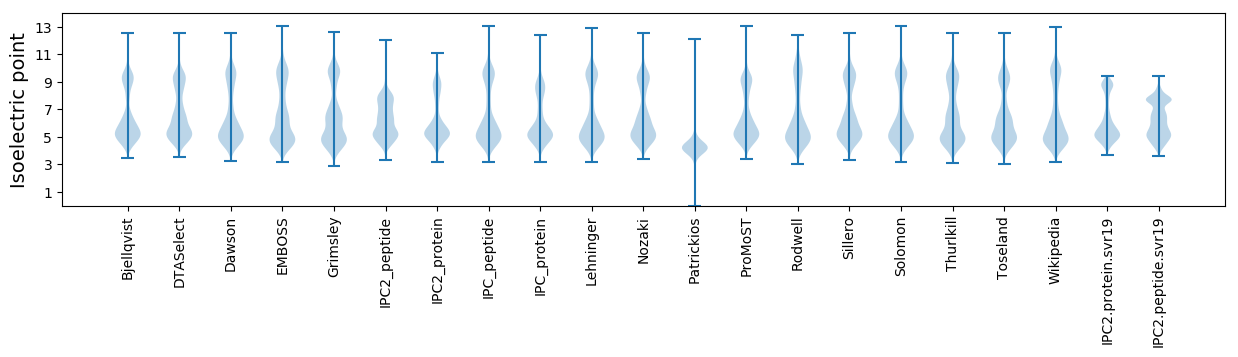
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S0I3B6|A0A3S0I3B6_9BACI HAMP domain-containing protein OS=Lysinibacillus telephonicus OX=1714840 GN=EKG35_04675 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.54QPKK8 pKa = 9.44KK9 pKa = 7.96RR10 pKa = 11.84KK11 pKa = 8.69HH12 pKa = 5.94SKK14 pKa = 8.54VHH16 pKa = 5.68GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.19NGRR28 pKa = 11.84KK29 pKa = 8.91VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.75VLSAA44 pKa = 4.05
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.54QPKK8 pKa = 9.44KK9 pKa = 7.96RR10 pKa = 11.84KK11 pKa = 8.69HH12 pKa = 5.94SKK14 pKa = 8.54VHH16 pKa = 5.68GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.19NGRR28 pKa = 11.84KK29 pKa = 8.91VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.75VLSAA44 pKa = 4.05
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1178354 |
14 |
1912 |
297.6 |
33.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.84 ± 0.044 | 0.715 ± 0.011 |
4.941 ± 0.031 | 7.608 ± 0.046 |
4.659 ± 0.039 | 6.603 ± 0.037 |
1.911 ± 0.015 | 8.648 ± 0.041 |
6.83 ± 0.039 | 9.737 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.563 ± 0.016 | 4.952 ± 0.034 |
3.438 ± 0.026 | 3.71 ± 0.027 |
3.72 ± 0.032 | 6.132 ± 0.035 |
5.474 ± 0.032 | 6.928 ± 0.032 |
0.965 ± 0.013 | 3.626 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
