
Wickerhamiella sorbophila
Taxonomy: cellular organisms; Eukaryota; Opisthokonta;
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
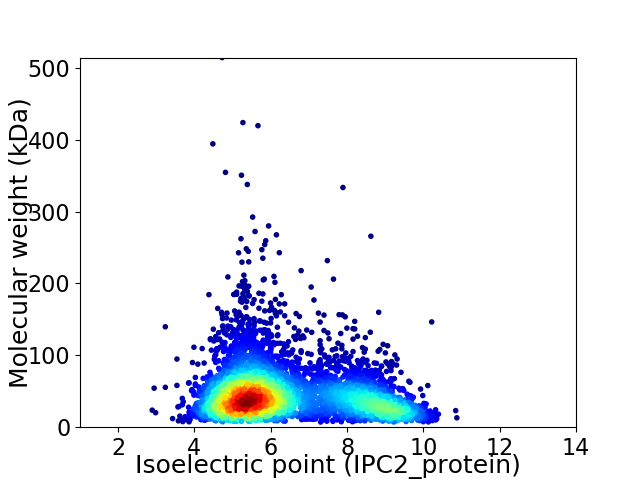
Virtual 2D-PAGE plot for 4707 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T0FL10|A0A2T0FL10_9ASCO Cytochrome b5 reductase 4 OS=Wickerhamiella sorbophila OX=45607 GN=B9G98_03289 PE=3 SV=1
MM1 pKa = 7.5LRR3 pKa = 11.84TIALFAGYY11 pKa = 10.54ALAQEE16 pKa = 4.26YY17 pKa = 10.24RR18 pKa = 11.84PVVPHH23 pKa = 6.34GRR25 pKa = 11.84EE26 pKa = 3.5EE27 pKa = 3.96QLRR30 pKa = 11.84QNAYY34 pKa = 9.14VDD36 pKa = 4.02VISTVVAPATQVFHH50 pKa = 6.17TTTDD54 pKa = 2.82VWVEE58 pKa = 3.84VYY60 pKa = 10.64PPVTTEE66 pKa = 3.85DD67 pKa = 3.36VTVFVPPPTHH77 pKa = 6.53TDD79 pKa = 3.51VYY81 pKa = 10.66WDD83 pKa = 4.19YY84 pKa = 12.03DD85 pKa = 3.92DD86 pKa = 6.75DD87 pKa = 7.12DD88 pKa = 6.98DD89 pKa = 6.02DD90 pKa = 5.28VYY92 pKa = 10.89EE93 pKa = 4.26TTVVVTALPGYY104 pKa = 8.55STTCEE109 pKa = 3.73TSTDD113 pKa = 4.01YY114 pKa = 11.3VDD116 pKa = 3.4KK117 pKa = 10.82TIVVPPAITSTSDD130 pKa = 2.95QSTTLPPVDD139 pKa = 3.51TTTGEE144 pKa = 4.22GSTTDD149 pKa = 3.62LTDD152 pKa = 3.18LTTFPITTATTEE164 pKa = 4.31STSEE168 pKa = 4.06SQTVHH173 pKa = 4.51TTVPSTVPTTDD184 pKa = 2.82STSGVTTEE192 pKa = 4.02STTDD196 pKa = 3.19VTTGTTTDD204 pKa = 3.47STSEE208 pKa = 4.17TTTNPATDD216 pKa = 3.83TTPSASTEE224 pKa = 4.23VEE226 pKa = 4.31TTTSNALPSTILPKK240 pKa = 10.66ASTPAPPAKK249 pKa = 9.51STPPPFTPTLPPQAEE264 pKa = 4.07NSDD267 pKa = 4.16IIPANGAYY275 pKa = 10.22NLQPLGSALYY285 pKa = 9.4FLLGIPLLFF294 pKa = 4.82
MM1 pKa = 7.5LRR3 pKa = 11.84TIALFAGYY11 pKa = 10.54ALAQEE16 pKa = 4.26YY17 pKa = 10.24RR18 pKa = 11.84PVVPHH23 pKa = 6.34GRR25 pKa = 11.84EE26 pKa = 3.5EE27 pKa = 3.96QLRR30 pKa = 11.84QNAYY34 pKa = 9.14VDD36 pKa = 4.02VISTVVAPATQVFHH50 pKa = 6.17TTTDD54 pKa = 2.82VWVEE58 pKa = 3.84VYY60 pKa = 10.64PPVTTEE66 pKa = 3.85DD67 pKa = 3.36VTVFVPPPTHH77 pKa = 6.53TDD79 pKa = 3.51VYY81 pKa = 10.66WDD83 pKa = 4.19YY84 pKa = 12.03DD85 pKa = 3.92DD86 pKa = 6.75DD87 pKa = 7.12DD88 pKa = 6.98DD89 pKa = 6.02DD90 pKa = 5.28VYY92 pKa = 10.89EE93 pKa = 4.26TTVVVTALPGYY104 pKa = 8.55STTCEE109 pKa = 3.73TSTDD113 pKa = 4.01YY114 pKa = 11.3VDD116 pKa = 3.4KK117 pKa = 10.82TIVVPPAITSTSDD130 pKa = 2.95QSTTLPPVDD139 pKa = 3.51TTTGEE144 pKa = 4.22GSTTDD149 pKa = 3.62LTDD152 pKa = 3.18LTTFPITTATTEE164 pKa = 4.31STSEE168 pKa = 4.06SQTVHH173 pKa = 4.51TTVPSTVPTTDD184 pKa = 2.82STSGVTTEE192 pKa = 4.02STTDD196 pKa = 3.19VTTGTTTDD204 pKa = 3.47STSEE208 pKa = 4.17TTTNPATDD216 pKa = 3.83TTPSASTEE224 pKa = 4.23VEE226 pKa = 4.31TTTSNALPSTILPKK240 pKa = 10.66ASTPAPPAKK249 pKa = 9.51STPPPFTPTLPPQAEE264 pKa = 4.07NSDD267 pKa = 4.16IIPANGAYY275 pKa = 10.22NLQPLGSALYY285 pKa = 9.4FLLGIPLLFF294 pKa = 4.82
Molecular weight: 31.18 kDa
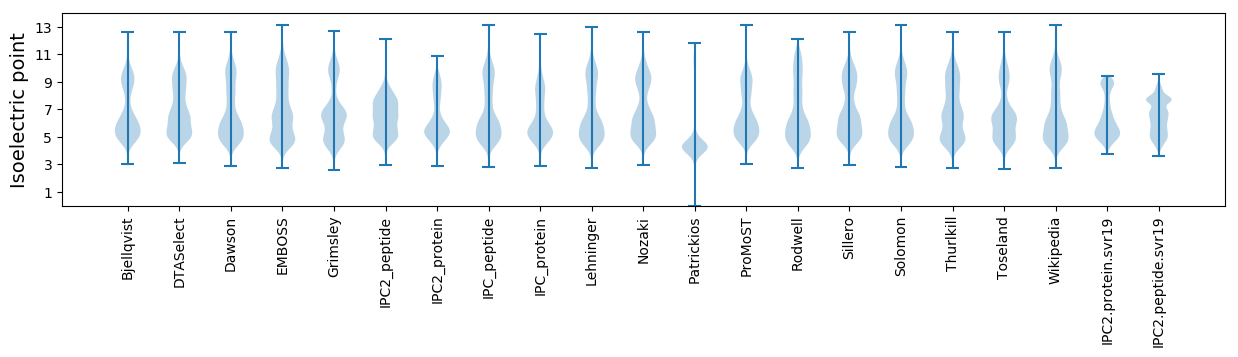
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T0FL07|A0A2T0FL07_9ASCO 40S ribosomal protein S12 OS=Wickerhamiella sorbophila OX=45607 GN=B9G98_03280 PE=3 SV=1
MM1 pKa = 7.38SAGARR6 pKa = 11.84SAGATRR12 pKa = 11.84SDD14 pKa = 3.65RR15 pKa = 11.84PRR17 pKa = 11.84LSARR21 pKa = 11.84ARR23 pKa = 11.84SAGATRR29 pKa = 11.84SDD31 pKa = 3.65RR32 pKa = 11.84PRR34 pKa = 11.84LSARR38 pKa = 11.84ARR40 pKa = 11.84SAGATRR46 pKa = 11.84SGRR49 pKa = 11.84PRR51 pKa = 11.84LGAGARR57 pKa = 11.84SAGASRR63 pKa = 11.84SARR66 pKa = 11.84PRR68 pKa = 11.84ASARR72 pKa = 11.84SRR74 pKa = 11.84GGDD77 pKa = 3.1RR78 pKa = 11.84SRR80 pKa = 11.84LSTRR84 pKa = 11.84ARR86 pKa = 11.84NAGATRR92 pKa = 11.84NDD94 pKa = 3.41RR95 pKa = 11.84AGLNTGARR103 pKa = 11.84SAGATRR109 pKa = 11.84SDD111 pKa = 3.6RR112 pKa = 11.84PRR114 pKa = 11.84ASARR118 pKa = 11.84SRR120 pKa = 11.84GGNRR124 pKa = 11.84SRR126 pKa = 11.84LHH128 pKa = 5.45TGARR132 pKa = 11.84SAGATGNNRR141 pKa = 11.84PRR143 pKa = 11.84LDD145 pKa = 3.11ARR147 pKa = 11.84ARR149 pKa = 11.84SAGTTRR155 pKa = 11.84NDD157 pKa = 3.56RR158 pKa = 11.84AGLNAGARR166 pKa = 11.84SAGATRR172 pKa = 11.84RR173 pKa = 11.84NRR175 pKa = 11.84PRR177 pKa = 11.84PSARR181 pKa = 11.84SRR183 pKa = 11.84GGDD186 pKa = 3.1RR187 pKa = 11.84SRR189 pKa = 11.84LSTRR193 pKa = 11.84TRR195 pKa = 11.84NAGTTRR201 pKa = 11.84SNRR204 pKa = 11.84SRR206 pKa = 11.84LNAGARR212 pKa = 11.84SAGDD216 pKa = 3.47TRR218 pKa = 11.84SDD220 pKa = 3.49RR221 pKa = 11.84PP222 pKa = 3.43
MM1 pKa = 7.38SAGARR6 pKa = 11.84SAGATRR12 pKa = 11.84SDD14 pKa = 3.65RR15 pKa = 11.84PRR17 pKa = 11.84LSARR21 pKa = 11.84ARR23 pKa = 11.84SAGATRR29 pKa = 11.84SDD31 pKa = 3.65RR32 pKa = 11.84PRR34 pKa = 11.84LSARR38 pKa = 11.84ARR40 pKa = 11.84SAGATRR46 pKa = 11.84SGRR49 pKa = 11.84PRR51 pKa = 11.84LGAGARR57 pKa = 11.84SAGASRR63 pKa = 11.84SARR66 pKa = 11.84PRR68 pKa = 11.84ASARR72 pKa = 11.84SRR74 pKa = 11.84GGDD77 pKa = 3.1RR78 pKa = 11.84SRR80 pKa = 11.84LSTRR84 pKa = 11.84ARR86 pKa = 11.84NAGATRR92 pKa = 11.84NDD94 pKa = 3.41RR95 pKa = 11.84AGLNTGARR103 pKa = 11.84SAGATRR109 pKa = 11.84SDD111 pKa = 3.6RR112 pKa = 11.84PRR114 pKa = 11.84ASARR118 pKa = 11.84SRR120 pKa = 11.84GGNRR124 pKa = 11.84SRR126 pKa = 11.84LHH128 pKa = 5.45TGARR132 pKa = 11.84SAGATGNNRR141 pKa = 11.84PRR143 pKa = 11.84LDD145 pKa = 3.11ARR147 pKa = 11.84ARR149 pKa = 11.84SAGTTRR155 pKa = 11.84NDD157 pKa = 3.56RR158 pKa = 11.84AGLNAGARR166 pKa = 11.84SAGATRR172 pKa = 11.84RR173 pKa = 11.84NRR175 pKa = 11.84PRR177 pKa = 11.84PSARR181 pKa = 11.84SRR183 pKa = 11.84GGDD186 pKa = 3.1RR187 pKa = 11.84SRR189 pKa = 11.84LSTRR193 pKa = 11.84TRR195 pKa = 11.84NAGTTRR201 pKa = 11.84SNRR204 pKa = 11.84SRR206 pKa = 11.84LNAGARR212 pKa = 11.84SAGDD216 pKa = 3.47TRR218 pKa = 11.84SDD220 pKa = 3.49RR221 pKa = 11.84PP222 pKa = 3.43
Molecular weight: 23.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2105241 |
65 |
4577 |
447.3 |
49.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.524 ± 0.042 | 1.16 ± 0.013 |
5.666 ± 0.027 | 6.514 ± 0.037 |
3.913 ± 0.029 | 5.977 ± 0.034 |
2.181 ± 0.014 | 5.098 ± 0.025 |
5.355 ± 0.032 | 9.583 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.21 ± 0.015 | 3.825 ± 0.017 |
5.411 ± 0.038 | 4.169 ± 0.028 |
5.587 ± 0.027 | 7.99 ± 0.047 |
5.624 ± 0.042 | 6.978 ± 0.026 |
1.224 ± 0.015 | 3.012 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
