
Drosophila guanche (Fruit fly)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Diptera; Brachycera; Muscomorpha; Eremoneura; Cyclorrhapha; Schizophora; Acalyptratae;
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
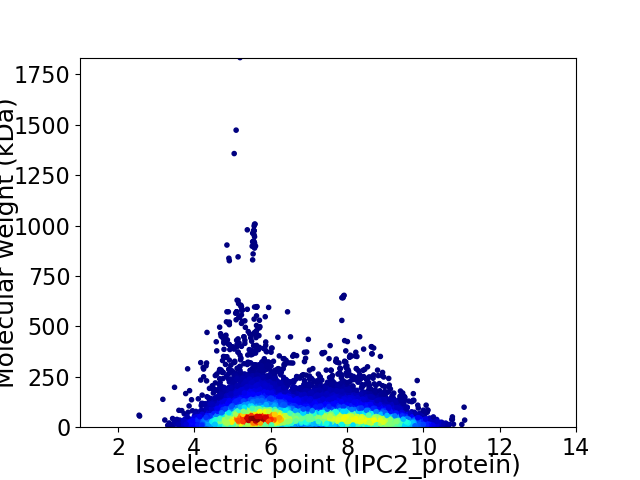
Virtual 2D-PAGE plot for 17569 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3B0KFM7|A0A3B0KFM7_DROGU Blast:Gamma-tubulin complex component 3 OS=Drosophila guanche OX=7266 GN=DGUA_6G014315 PE=4 SV=1
MM1 pKa = 7.43KK2 pKa = 10.19LAFIFMSVQLMLLATSLNARR22 pKa = 11.84RR23 pKa = 11.84CIRR26 pKa = 11.84PTTTTTSTPKK36 pKa = 8.49TTDD39 pKa = 3.83DD40 pKa = 3.94EE41 pKa = 4.89PSDD44 pKa = 3.97EE45 pKa = 4.75SSDD48 pKa = 3.59SSGEE52 pKa = 3.89ITNAPSDD59 pKa = 3.93PTGEE63 pKa = 4.6TTGSTPDD70 pKa = 3.69PTSKK74 pKa = 9.38PTDD77 pKa = 3.69APSDD81 pKa = 3.73PTGEE85 pKa = 4.53TTGSPSDD92 pKa = 3.69PTIEE96 pKa = 4.22PTDD99 pKa = 3.98APSDD103 pKa = 3.91PTGKK107 pKa = 9.88PSDD110 pKa = 4.37APSDD114 pKa = 3.94PPGEE118 pKa = 4.48TTGPPSDD125 pKa = 3.87ATDD128 pKa = 3.61MPTDD132 pKa = 3.8APSNPTGKK140 pKa = 8.42PTDD143 pKa = 4.09APSDD147 pKa = 3.92PPGEE151 pKa = 4.38TTNAAPDD158 pKa = 3.91PSRR161 pKa = 11.84EE162 pKa = 3.81TSAAPSDD169 pKa = 3.67PTDD172 pKa = 3.56ASGVPADD179 pKa = 4.09TTQTTTPPPAIDD191 pKa = 4.57SDD193 pKa = 4.5AACQAHH199 pKa = 6.9PGVHH203 pKa = 6.04YY204 pKa = 10.63LPNPYY209 pKa = 9.86DD210 pKa = 3.37CHH212 pKa = 8.95KK213 pKa = 10.86YY214 pKa = 9.15IQCEE218 pKa = 4.19GNVGHH223 pKa = 7.3IHH225 pKa = 6.3TCPADD230 pKa = 4.1LYY232 pKa = 9.07WNSIALTCDD241 pKa = 3.52TVCQQ245 pKa = 3.72
MM1 pKa = 7.43KK2 pKa = 10.19LAFIFMSVQLMLLATSLNARR22 pKa = 11.84RR23 pKa = 11.84CIRR26 pKa = 11.84PTTTTTSTPKK36 pKa = 8.49TTDD39 pKa = 3.83DD40 pKa = 3.94EE41 pKa = 4.89PSDD44 pKa = 3.97EE45 pKa = 4.75SSDD48 pKa = 3.59SSGEE52 pKa = 3.89ITNAPSDD59 pKa = 3.93PTGEE63 pKa = 4.6TTGSTPDD70 pKa = 3.69PTSKK74 pKa = 9.38PTDD77 pKa = 3.69APSDD81 pKa = 3.73PTGEE85 pKa = 4.53TTGSPSDD92 pKa = 3.69PTIEE96 pKa = 4.22PTDD99 pKa = 3.98APSDD103 pKa = 3.91PTGKK107 pKa = 9.88PSDD110 pKa = 4.37APSDD114 pKa = 3.94PPGEE118 pKa = 4.48TTGPPSDD125 pKa = 3.87ATDD128 pKa = 3.61MPTDD132 pKa = 3.8APSNPTGKK140 pKa = 8.42PTDD143 pKa = 4.09APSDD147 pKa = 3.92PPGEE151 pKa = 4.38TTNAAPDD158 pKa = 3.91PSRR161 pKa = 11.84EE162 pKa = 3.81TSAAPSDD169 pKa = 3.67PTDD172 pKa = 3.56ASGVPADD179 pKa = 4.09TTQTTTPPPAIDD191 pKa = 4.57SDD193 pKa = 4.5AACQAHH199 pKa = 6.9PGVHH203 pKa = 6.04YY204 pKa = 10.63LPNPYY209 pKa = 9.86DD210 pKa = 3.37CHH212 pKa = 8.95KK213 pKa = 10.86YY214 pKa = 9.15IQCEE218 pKa = 4.19GNVGHH223 pKa = 7.3IHH225 pKa = 6.3TCPADD230 pKa = 4.1LYY232 pKa = 9.07WNSIALTCDD241 pKa = 3.52TVCQQ245 pKa = 3.72
Molecular weight: 25.18 kDa
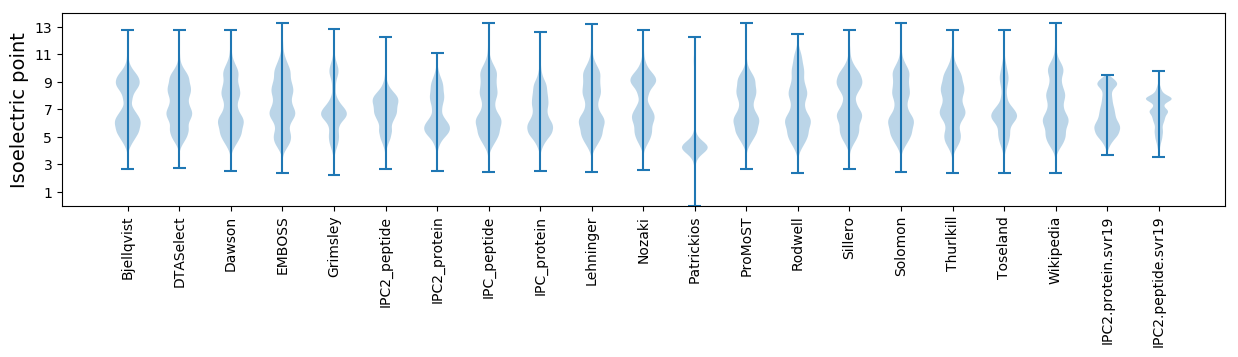
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3B0JPY2|A0A3B0JPY2_DROGU Isoform of A0A3B0JPP2 Blast:Sorbitol dehydrogenase OS=Drosophila guanche OX=7266 GN=DGUA_6G016714 PE=3 SV=1
MM1 pKa = 7.64RR2 pKa = 11.84ADD4 pKa = 5.5LIANSNLTRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84SGSSTRR21 pKa = 11.84RR22 pKa = 11.84SSSAVGGLRR31 pKa = 11.84QRR33 pKa = 11.84LGNRR37 pKa = 11.84RR38 pKa = 11.84PSVGGASEE46 pKa = 3.83RR47 pKa = 11.84VEE49 pKa = 3.72RR50 pKa = 11.84HH51 pKa = 3.59QRR53 pKa = 11.84QLIKK57 pKa = 10.34QQQQQYY63 pKa = 10.35HH64 pKa = 5.83LQQTRR69 pKa = 11.84GRR71 pKa = 11.84SRR73 pKa = 11.84SRR75 pKa = 11.84SRR77 pKa = 11.84SRR79 pKa = 11.84LPQVNARR86 pKa = 11.84IDD88 pKa = 3.43RR89 pKa = 11.84SRR91 pKa = 11.84SQSRR95 pKa = 11.84GRR97 pKa = 11.84SAAPQFNRR105 pKa = 11.84TRR107 pKa = 11.84SQSRR111 pKa = 11.84TRR113 pKa = 11.84GGRR116 pKa = 11.84DD117 pKa = 2.92RR118 pKa = 11.84RR119 pKa = 11.84PQSGTRR125 pKa = 11.84AGQQTPRR132 pKa = 11.84VSVKK136 pKa = 9.64QRR138 pKa = 11.84LGVRR142 pKa = 11.84PGQGRR147 pKa = 11.84GGAGARR153 pKa = 11.84TRR155 pKa = 11.84AGANQAKK162 pKa = 8.27NQRR165 pKa = 11.84RR166 pKa = 11.84APSKK170 pKa = 10.39LRR172 pKa = 11.84GVAGGRR178 pKa = 11.84VEE180 pKa = 4.29KK181 pKa = 10.56RR182 pKa = 11.84RR183 pKa = 11.84NANPTQKK190 pKa = 10.52GVSASLAGRR199 pKa = 11.84RR200 pKa = 11.84GRR202 pKa = 11.84PRR204 pKa = 11.84GRR206 pKa = 11.84SVARR210 pKa = 11.84AAAAAGGGAASGSNGASTRR229 pKa = 11.84RR230 pKa = 11.84SRR232 pKa = 11.84SRR234 pKa = 11.84NRR236 pKa = 11.84PAAAAVAAAAATPGRR251 pKa = 11.84ANNTRR256 pKa = 11.84NRR258 pKa = 11.84RR259 pKa = 11.84GRR261 pKa = 11.84SAGAVKK267 pKa = 10.68GSGAKK272 pKa = 9.59GAAANKK278 pKa = 10.0NGNKK282 pKa = 10.04SNASKK287 pKa = 11.2GKK289 pKa = 7.19TNKK292 pKa = 9.21GRR294 pKa = 11.84PGGGGKK300 pKa = 9.46PGPQQQQARR309 pKa = 11.84RR310 pKa = 11.84GRR312 pKa = 11.84SRR314 pKa = 11.84TRR316 pKa = 11.84AGRR319 pKa = 11.84QAPGKK324 pKa = 10.14SVTSHH329 pKa = 6.18EE330 pKa = 4.24AHH332 pKa = 6.51SS333 pKa = 3.73
MM1 pKa = 7.64RR2 pKa = 11.84ADD4 pKa = 5.5LIANSNLTRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84SGSSTRR21 pKa = 11.84RR22 pKa = 11.84SSSAVGGLRR31 pKa = 11.84QRR33 pKa = 11.84LGNRR37 pKa = 11.84RR38 pKa = 11.84PSVGGASEE46 pKa = 3.83RR47 pKa = 11.84VEE49 pKa = 3.72RR50 pKa = 11.84HH51 pKa = 3.59QRR53 pKa = 11.84QLIKK57 pKa = 10.34QQQQQYY63 pKa = 10.35HH64 pKa = 5.83LQQTRR69 pKa = 11.84GRR71 pKa = 11.84SRR73 pKa = 11.84SRR75 pKa = 11.84SRR77 pKa = 11.84SRR79 pKa = 11.84LPQVNARR86 pKa = 11.84IDD88 pKa = 3.43RR89 pKa = 11.84SRR91 pKa = 11.84SQSRR95 pKa = 11.84GRR97 pKa = 11.84SAAPQFNRR105 pKa = 11.84TRR107 pKa = 11.84SQSRR111 pKa = 11.84TRR113 pKa = 11.84GGRR116 pKa = 11.84DD117 pKa = 2.92RR118 pKa = 11.84RR119 pKa = 11.84PQSGTRR125 pKa = 11.84AGQQTPRR132 pKa = 11.84VSVKK136 pKa = 9.64QRR138 pKa = 11.84LGVRR142 pKa = 11.84PGQGRR147 pKa = 11.84GGAGARR153 pKa = 11.84TRR155 pKa = 11.84AGANQAKK162 pKa = 8.27NQRR165 pKa = 11.84RR166 pKa = 11.84APSKK170 pKa = 10.39LRR172 pKa = 11.84GVAGGRR178 pKa = 11.84VEE180 pKa = 4.29KK181 pKa = 10.56RR182 pKa = 11.84RR183 pKa = 11.84NANPTQKK190 pKa = 10.52GVSASLAGRR199 pKa = 11.84RR200 pKa = 11.84GRR202 pKa = 11.84PRR204 pKa = 11.84GRR206 pKa = 11.84SVARR210 pKa = 11.84AAAAAGGGAASGSNGASTRR229 pKa = 11.84RR230 pKa = 11.84SRR232 pKa = 11.84SRR234 pKa = 11.84NRR236 pKa = 11.84PAAAAVAAAAATPGRR251 pKa = 11.84ANNTRR256 pKa = 11.84NRR258 pKa = 11.84RR259 pKa = 11.84GRR261 pKa = 11.84SAGAVKK267 pKa = 10.68GSGAKK272 pKa = 9.59GAAANKK278 pKa = 10.0NGNKK282 pKa = 10.04SNASKK287 pKa = 11.2GKK289 pKa = 7.19TNKK292 pKa = 9.21GRR294 pKa = 11.84PGGGGKK300 pKa = 9.46PGPQQQQARR309 pKa = 11.84RR310 pKa = 11.84GRR312 pKa = 11.84SRR314 pKa = 11.84TRR316 pKa = 11.84AGRR319 pKa = 11.84QAPGKK324 pKa = 10.14SVTSHH329 pKa = 6.18EE330 pKa = 4.24AHH332 pKa = 6.51SS333 pKa = 3.73
Molecular weight: 35.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
10893509 |
14 |
17223 |
620.0 |
68.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.811 ± 0.026 | 1.913 ± 0.027 |
5.114 ± 0.017 | 6.386 ± 0.024 |
3.355 ± 0.014 | 6.18 ± 0.023 |
2.73 ± 0.011 | 4.696 ± 0.016 |
5.381 ± 0.026 | 8.987 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.355 ± 0.01 | 4.602 ± 0.013 |
5.611 ± 0.029 | 5.466 ± 0.029 |
5.484 ± 0.015 | 8.428 ± 0.027 |
5.824 ± 0.02 | 5.813 ± 0.015 |
0.971 ± 0.007 | 2.895 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
