
Helminthosporium victoriae virus-190S (Hv190SV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins
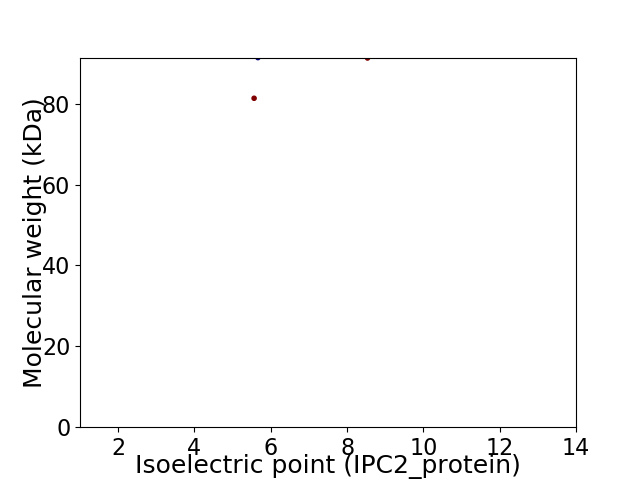
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
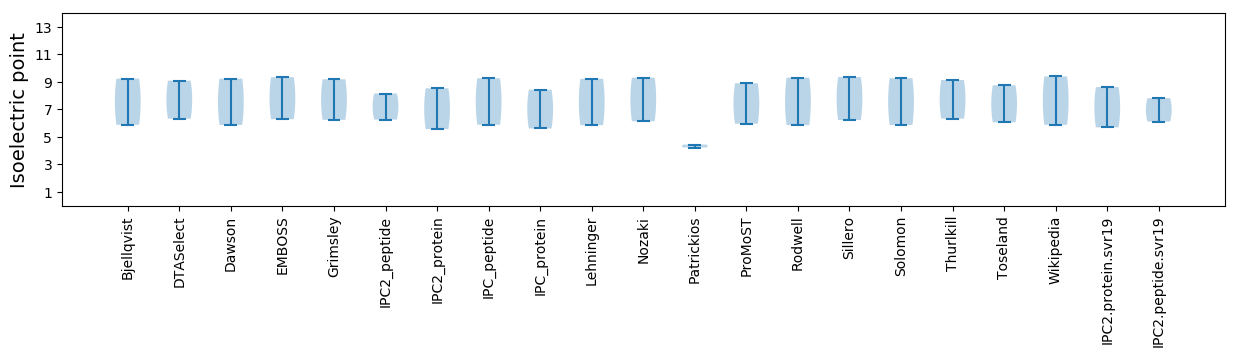
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|O57044|RDRP_HV19S Probable RNA-directed RNA polymerase OS=Helminthosporium victoriae virus-190S OX=45237 PE=3 SV=2
MM1 pKa = 7.23SHH3 pKa = 4.89TTITNFLAGVIARR16 pKa = 11.84PQGGNITSDD25 pKa = 3.19EE26 pKa = 4.06TFRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 8.57RR32 pKa = 11.84TIVRR36 pKa = 11.84TSATIGGNEE45 pKa = 4.31DD46 pKa = 3.25SRR48 pKa = 11.84TTSIFHH54 pKa = 6.62EE55 pKa = 4.35IGRR58 pKa = 11.84AVNTKK63 pKa = 9.83GKK65 pKa = 9.31ALAVAGMEE73 pKa = 4.16APLVEE78 pKa = 4.61ASYY81 pKa = 8.44PTNAVLVEE89 pKa = 4.21DD90 pKa = 5.6FIGLAKK96 pKa = 10.54KK97 pKa = 8.47YY98 pKa = 8.7TNFSATFEE106 pKa = 4.19YY107 pKa = 10.9SSLAGVVEE115 pKa = 4.37RR116 pKa = 11.84LARR119 pKa = 11.84GLAACSVFEE128 pKa = 5.12DD129 pKa = 3.95VTSTDD134 pKa = 3.2LRR136 pKa = 11.84GNNPLAVHH144 pKa = 7.08ALATYY149 pKa = 9.45DD150 pKa = 3.83GPVNSLTSAVFIPRR164 pKa = 11.84LVNNALTGDD173 pKa = 4.13VFAVLCNCVAGEE185 pKa = 4.33GGTVVTDD192 pKa = 4.36TIEE195 pKa = 4.56LDD197 pKa = 3.3ANTRR201 pKa = 11.84QPIVPEE207 pKa = 4.02VGPLGVPGAIVDD219 pKa = 3.99ALRR222 pKa = 11.84LLGSNMIASDD232 pKa = 3.74QGPLFALALTRR243 pKa = 11.84GIHH246 pKa = 5.8RR247 pKa = 11.84VLSVVGHH254 pKa = 5.44TDD256 pKa = 2.38EE257 pKa = 5.21GGIVRR262 pKa = 11.84DD263 pKa = 3.84LLRR266 pKa = 11.84CGGFGLPFGGIHH278 pKa = 6.37YY279 pKa = 10.51GLEE282 pKa = 4.34EE283 pKa = 4.27YY284 pKa = 10.7SGLPALQFNSAAATAAYY301 pKa = 10.28VDD303 pKa = 5.29GIALVTAAVVAHH315 pKa = 7.11ADD317 pKa = 3.11PGEE320 pKa = 4.32RR321 pKa = 11.84YY322 pKa = 9.71NGEE325 pKa = 3.78WFPTFFDD332 pKa = 4.04GTTHH336 pKa = 7.16ADD338 pKa = 3.09TMRR341 pKa = 11.84RR342 pKa = 11.84SGDD345 pKa = 3.61STEE348 pKa = 4.21GTAAMADD355 pKa = 3.83RR356 pKa = 11.84NRR358 pKa = 11.84AQLLARR364 pKa = 11.84QQLFWRR370 pKa = 11.84PYY372 pKa = 7.52ITALGACFSTAGDD385 pKa = 3.2ISVAEE390 pKa = 4.33RR391 pKa = 11.84FQCAASHH398 pKa = 6.46SLGADD403 pKa = 3.36PRR405 pKa = 11.84HH406 pKa = 5.85LRR408 pKa = 11.84LPSVAPYY415 pKa = 9.79FWIEE419 pKa = 3.72PTGLIPHH426 pKa = 7.31DD427 pKa = 3.86FLGSVAEE434 pKa = 4.25EE435 pKa = 4.06EE436 pKa = 5.1GFASYY441 pKa = 10.41CWRR444 pKa = 11.84DD445 pKa = 3.56TTRR448 pKa = 11.84TRR450 pKa = 11.84PAWDD454 pKa = 3.85SIVLSGARR462 pKa = 11.84DD463 pKa = 3.77TTFSAYY469 pKa = 10.01HH470 pKa = 6.39IRR472 pKa = 11.84MKK474 pKa = 10.49GARR477 pKa = 11.84TAWFLAHH484 pKa = 7.03WLGHH488 pKa = 5.84PEE490 pKa = 4.03NGLGATRR497 pKa = 11.84VRR499 pKa = 11.84QLDD502 pKa = 3.91PNAVLHH508 pKa = 6.43PGPCEE513 pKa = 3.74GNEE516 pKa = 3.89QVRR519 pKa = 11.84DD520 pKa = 3.53RR521 pKa = 11.84VEE523 pKa = 4.62ADD525 pKa = 4.07LPLTDD530 pKa = 3.83YY531 pKa = 10.94LWRR534 pKa = 11.84RR535 pKa = 11.84GQSPFPAAGEE545 pKa = 4.04LLNLTSEE552 pKa = 3.76WGILFRR558 pKa = 11.84HH559 pKa = 5.35VTFTDD564 pKa = 4.01DD565 pKa = 3.5GDD567 pKa = 4.51LNPEE571 pKa = 4.29HH572 pKa = 6.96LPAAHH577 pKa = 6.69EE578 pKa = 4.21MADD581 pKa = 3.56TTVTMTVGRR590 pKa = 11.84PIGIAPGRR598 pKa = 11.84SNAGDD603 pKa = 3.45NQARR607 pKa = 11.84RR608 pKa = 11.84ARR610 pKa = 11.84TRR612 pKa = 11.84ASVEE616 pKa = 3.42LSAASRR622 pKa = 11.84RR623 pKa = 11.84ARR625 pKa = 11.84VFGRR629 pKa = 11.84PDD631 pKa = 3.24VGEE634 pKa = 4.23MPTLTSAPAPIPASPAYY651 pKa = 10.01DD652 pKa = 3.34GNRR655 pKa = 11.84GGEE658 pKa = 4.05AGGVTGRR665 pKa = 11.84GNNRR669 pKa = 11.84SAAPGHH675 pKa = 6.16ASWSEE680 pKa = 3.84RR681 pKa = 11.84QADD684 pKa = 3.98GVPVNVTPHH693 pKa = 6.25HH694 pKa = 5.92NALRR698 pKa = 11.84APPFPRR704 pKa = 11.84QQGALGGGGNVPLPPAPGAAPPPPPGPPNGPPAGPPPSDD743 pKa = 4.45DD744 pKa = 3.71GSSNPAAPVPTAIHH758 pKa = 6.69APPAAAQADD767 pKa = 4.26RR768 pKa = 11.84AEE770 pKa = 4.4GQQ772 pKa = 3.15
MM1 pKa = 7.23SHH3 pKa = 4.89TTITNFLAGVIARR16 pKa = 11.84PQGGNITSDD25 pKa = 3.19EE26 pKa = 4.06TFRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 8.57RR32 pKa = 11.84TIVRR36 pKa = 11.84TSATIGGNEE45 pKa = 4.31DD46 pKa = 3.25SRR48 pKa = 11.84TTSIFHH54 pKa = 6.62EE55 pKa = 4.35IGRR58 pKa = 11.84AVNTKK63 pKa = 9.83GKK65 pKa = 9.31ALAVAGMEE73 pKa = 4.16APLVEE78 pKa = 4.61ASYY81 pKa = 8.44PTNAVLVEE89 pKa = 4.21DD90 pKa = 5.6FIGLAKK96 pKa = 10.54KK97 pKa = 8.47YY98 pKa = 8.7TNFSATFEE106 pKa = 4.19YY107 pKa = 10.9SSLAGVVEE115 pKa = 4.37RR116 pKa = 11.84LARR119 pKa = 11.84GLAACSVFEE128 pKa = 5.12DD129 pKa = 3.95VTSTDD134 pKa = 3.2LRR136 pKa = 11.84GNNPLAVHH144 pKa = 7.08ALATYY149 pKa = 9.45DD150 pKa = 3.83GPVNSLTSAVFIPRR164 pKa = 11.84LVNNALTGDD173 pKa = 4.13VFAVLCNCVAGEE185 pKa = 4.33GGTVVTDD192 pKa = 4.36TIEE195 pKa = 4.56LDD197 pKa = 3.3ANTRR201 pKa = 11.84QPIVPEE207 pKa = 4.02VGPLGVPGAIVDD219 pKa = 3.99ALRR222 pKa = 11.84LLGSNMIASDD232 pKa = 3.74QGPLFALALTRR243 pKa = 11.84GIHH246 pKa = 5.8RR247 pKa = 11.84VLSVVGHH254 pKa = 5.44TDD256 pKa = 2.38EE257 pKa = 5.21GGIVRR262 pKa = 11.84DD263 pKa = 3.84LLRR266 pKa = 11.84CGGFGLPFGGIHH278 pKa = 6.37YY279 pKa = 10.51GLEE282 pKa = 4.34EE283 pKa = 4.27YY284 pKa = 10.7SGLPALQFNSAAATAAYY301 pKa = 10.28VDD303 pKa = 5.29GIALVTAAVVAHH315 pKa = 7.11ADD317 pKa = 3.11PGEE320 pKa = 4.32RR321 pKa = 11.84YY322 pKa = 9.71NGEE325 pKa = 3.78WFPTFFDD332 pKa = 4.04GTTHH336 pKa = 7.16ADD338 pKa = 3.09TMRR341 pKa = 11.84RR342 pKa = 11.84SGDD345 pKa = 3.61STEE348 pKa = 4.21GTAAMADD355 pKa = 3.83RR356 pKa = 11.84NRR358 pKa = 11.84AQLLARR364 pKa = 11.84QQLFWRR370 pKa = 11.84PYY372 pKa = 7.52ITALGACFSTAGDD385 pKa = 3.2ISVAEE390 pKa = 4.33RR391 pKa = 11.84FQCAASHH398 pKa = 6.46SLGADD403 pKa = 3.36PRR405 pKa = 11.84HH406 pKa = 5.85LRR408 pKa = 11.84LPSVAPYY415 pKa = 9.79FWIEE419 pKa = 3.72PTGLIPHH426 pKa = 7.31DD427 pKa = 3.86FLGSVAEE434 pKa = 4.25EE435 pKa = 4.06EE436 pKa = 5.1GFASYY441 pKa = 10.41CWRR444 pKa = 11.84DD445 pKa = 3.56TTRR448 pKa = 11.84TRR450 pKa = 11.84PAWDD454 pKa = 3.85SIVLSGARR462 pKa = 11.84DD463 pKa = 3.77TTFSAYY469 pKa = 10.01HH470 pKa = 6.39IRR472 pKa = 11.84MKK474 pKa = 10.49GARR477 pKa = 11.84TAWFLAHH484 pKa = 7.03WLGHH488 pKa = 5.84PEE490 pKa = 4.03NGLGATRR497 pKa = 11.84VRR499 pKa = 11.84QLDD502 pKa = 3.91PNAVLHH508 pKa = 6.43PGPCEE513 pKa = 3.74GNEE516 pKa = 3.89QVRR519 pKa = 11.84DD520 pKa = 3.53RR521 pKa = 11.84VEE523 pKa = 4.62ADD525 pKa = 4.07LPLTDD530 pKa = 3.83YY531 pKa = 10.94LWRR534 pKa = 11.84RR535 pKa = 11.84GQSPFPAAGEE545 pKa = 4.04LLNLTSEE552 pKa = 3.76WGILFRR558 pKa = 11.84HH559 pKa = 5.35VTFTDD564 pKa = 4.01DD565 pKa = 3.5GDD567 pKa = 4.51LNPEE571 pKa = 4.29HH572 pKa = 6.96LPAAHH577 pKa = 6.69EE578 pKa = 4.21MADD581 pKa = 3.56TTVTMTVGRR590 pKa = 11.84PIGIAPGRR598 pKa = 11.84SNAGDD603 pKa = 3.45NQARR607 pKa = 11.84RR608 pKa = 11.84ARR610 pKa = 11.84TRR612 pKa = 11.84ASVEE616 pKa = 3.42LSAASRR622 pKa = 11.84RR623 pKa = 11.84ARR625 pKa = 11.84VFGRR629 pKa = 11.84PDD631 pKa = 3.24VGEE634 pKa = 4.23MPTLTSAPAPIPASPAYY651 pKa = 10.01DD652 pKa = 3.34GNRR655 pKa = 11.84GGEE658 pKa = 4.05AGGVTGRR665 pKa = 11.84GNNRR669 pKa = 11.84SAAPGHH675 pKa = 6.16ASWSEE680 pKa = 3.84RR681 pKa = 11.84QADD684 pKa = 3.98GVPVNVTPHH693 pKa = 6.25HH694 pKa = 5.92NALRR698 pKa = 11.84APPFPRR704 pKa = 11.84QQGALGGGGNVPLPPAPGAAPPPPPGPPNGPPAGPPPSDD743 pKa = 4.45DD744 pKa = 3.71GSSNPAAPVPTAIHH758 pKa = 6.69APPAAAQADD767 pKa = 4.26RR768 pKa = 11.84AEE770 pKa = 4.4GQQ772 pKa = 3.15
Molecular weight: 81.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|O57044|RDRP_HV19S Probable RNA-directed RNA polymerase OS=Helminthosporium victoriae virus-190S OX=45237 PE=3 SV=2
MM1 pKa = 7.96SDD3 pKa = 3.1PQEE6 pKa = 3.8RR7 pKa = 11.84SKK9 pKa = 11.34AYY11 pKa = 9.94GLLGEE16 pKa = 4.57RR17 pKa = 11.84LYY19 pKa = 11.36AVASANSHH27 pKa = 5.34MLAGYY32 pKa = 10.42DD33 pKa = 3.81SLDD36 pKa = 3.22FTARR40 pKa = 11.84LVRR43 pKa = 11.84LTGEE47 pKa = 3.93ATALKK52 pKa = 10.39AVDD55 pKa = 4.32PLLPCAVSLLFMDD68 pKa = 6.31FPLQLPCTPEE78 pKa = 3.42EE79 pKa = 3.92TLRR82 pKa = 11.84LVRR85 pKa = 11.84RR86 pKa = 11.84AYY88 pKa = 10.41DD89 pKa = 3.57PNTLEE94 pKa = 4.04EE95 pKa = 4.09VDD97 pKa = 3.89YY98 pKa = 10.23STLSGYY104 pKa = 7.97ATQFARR110 pKa = 11.84VKK112 pKa = 9.46GQRR115 pKa = 11.84GRR117 pKa = 11.84WRR119 pKa = 11.84HH120 pKa = 5.7LGHH123 pKa = 7.31LVCNDD128 pKa = 2.56KK129 pKa = 11.1AFRR132 pKa = 11.84EE133 pKa = 4.38RR134 pKa = 11.84YY135 pKa = 8.97FPKK138 pKa = 10.22KK139 pKa = 9.47KK140 pKa = 9.78HH141 pKa = 5.25AAAAIKK147 pKa = 9.65TNIRR151 pKa = 11.84LGPLARR157 pKa = 11.84AWAARR162 pKa = 11.84YY163 pKa = 9.14GLAALGSHH171 pKa = 7.08LAYY174 pKa = 9.66MVGMPNDD181 pKa = 3.88RR182 pKa = 11.84ACATLLLAQTYY193 pKa = 8.78KK194 pKa = 11.02ARR196 pKa = 11.84FGSEE200 pKa = 3.6GVAWAIASVRR210 pKa = 11.84QPEE213 pKa = 4.2NAKK216 pKa = 10.47GLSNALKK223 pKa = 10.57ALGSNTSEE231 pKa = 4.0PGALFVEE238 pKa = 4.86ANTLQGRR245 pKa = 11.84YY246 pKa = 9.91DD247 pKa = 3.37RR248 pKa = 11.84TLDD251 pKa = 3.26MDD253 pKa = 4.12HH254 pKa = 6.69EE255 pKa = 4.73VEE257 pKa = 4.71SRR259 pKa = 11.84CSPAAIADD267 pKa = 3.62QVIPYY272 pKa = 9.05TDD274 pKa = 3.53EE275 pKa = 4.61LGACIDD281 pKa = 4.91FILDD285 pKa = 3.61TEE287 pKa = 4.73LGGDD291 pKa = 3.88TIEE294 pKa = 6.26LPDD297 pKa = 4.05EE298 pKa = 4.62DD299 pKa = 4.5EE300 pKa = 4.09WWTSRR305 pKa = 11.84WLWCVNGSQNALSDD319 pKa = 3.44KK320 pKa = 11.21ALGIKK325 pKa = 9.88NKK327 pKa = 10.12SGQRR331 pKa = 11.84YY332 pKa = 8.22RR333 pKa = 11.84RR334 pKa = 11.84MAAEE338 pKa = 4.01EE339 pKa = 4.37VNNNPVPAWNGHH351 pKa = 4.63TSVSPSVKK359 pKa = 10.42LEE361 pKa = 3.85NGKK364 pKa = 9.89DD365 pKa = 3.23RR366 pKa = 11.84AIFACDD372 pKa = 3.05TRR374 pKa = 11.84SYY376 pKa = 11.07FAFTYY381 pKa = 9.55WLTPIEE387 pKa = 4.46KK388 pKa = 8.58KK389 pKa = 9.44WRR391 pKa = 11.84GARR394 pKa = 11.84VILNPGEE401 pKa = 4.14GGLYY405 pKa = 8.56GTARR409 pKa = 11.84RR410 pKa = 11.84IRR412 pKa = 11.84GSQTSGGVNLMLDD425 pKa = 3.53YY426 pKa = 11.59DD427 pKa = 4.27NFNSQHH433 pKa = 6.26SNEE436 pKa = 4.15TMAALYY442 pKa = 9.4EE443 pKa = 4.29KK444 pKa = 10.73ALSRR448 pKa = 11.84TNAPAYY454 pKa = 9.99LKK456 pKa = 10.51KK457 pKa = 10.55AVAASVEE464 pKa = 4.3STYY467 pKa = 10.92IHH469 pKa = 6.15YY470 pKa = 10.38KK471 pKa = 9.92GRR473 pKa = 11.84DD474 pKa = 3.08RR475 pKa = 11.84HH476 pKa = 5.76VLGTLMSGHH485 pKa = 7.19RR486 pKa = 11.84ATTFTNSVLNAAYY499 pKa = 8.95ICYY502 pKa = 10.2AVGIPAFKK510 pKa = 10.75RR511 pKa = 11.84MISLHH516 pKa = 6.61AGDD519 pKa = 5.45DD520 pKa = 3.79VYY522 pKa = 11.72LRR524 pKa = 11.84LPTLADD530 pKa = 3.61CATTLNNTKK539 pKa = 10.38RR540 pKa = 11.84VGCRR544 pKa = 11.84MNPTKK549 pKa = 10.55QSIGYY554 pKa = 7.82TGAEE558 pKa = 4.29FLRR561 pKa = 11.84LGINKK566 pKa = 9.61SYY568 pKa = 10.93AIGYY572 pKa = 7.5LCRR575 pKa = 11.84AIASLVSGSWTSLDD589 pKa = 3.4EE590 pKa = 4.23LQPLNALNGAIVQTRR605 pKa = 11.84SCLNRR610 pKa = 11.84GAATGLPEE618 pKa = 5.6LISASFVGLRR628 pKa = 11.84GFKK631 pKa = 10.4RR632 pKa = 11.84RR633 pKa = 11.84DD634 pKa = 3.39LLEE637 pKa = 4.51LLTGVATIKK646 pKa = 10.27PGPVYY651 pKa = 10.29TSSCVIRR658 pKa = 11.84EE659 pKa = 4.17YY660 pKa = 11.04VVEE663 pKa = 4.28QPPPPQFDD671 pKa = 3.89VPPGAGMHH679 pKa = 5.84ATMSYY684 pKa = 10.24LARR687 pKa = 11.84HH688 pKa = 4.9TTLVEE693 pKa = 3.96AQALEE698 pKa = 4.08IARR701 pKa = 11.84PAIKK705 pKa = 10.43SLMLSSSYY713 pKa = 11.11GKK715 pKa = 10.27AGPGAQTRR723 pKa = 11.84PHH725 pKa = 7.54VPMPKK730 pKa = 9.85LRR732 pKa = 11.84RR733 pKa = 11.84QPPRR737 pKa = 11.84VAVGFSMAHH746 pKa = 6.23EE747 pKa = 4.02LTGRR751 pKa = 11.84GVKK754 pKa = 9.67EE755 pKa = 4.3GCLSGHH761 pKa = 6.32PLLRR765 pKa = 11.84LFEE768 pKa = 4.06QRR770 pKa = 11.84LTDD773 pKa = 4.05DD774 pKa = 4.21DD775 pKa = 4.78LRR777 pKa = 11.84ALVALVGGNTSAKK790 pKa = 10.14DD791 pKa = 3.11IRR793 pKa = 11.84AEE795 pKa = 3.89AFGAEE800 pKa = 4.42SSSSTIMGILPHH812 pKa = 6.92SDD814 pKa = 2.3ASNYY818 pKa = 9.62CKK820 pKa = 9.86RR821 pKa = 11.84TRR823 pKa = 11.84NGNIIVPYY831 pKa = 9.8HH832 pKa = 5.86IRR834 pKa = 11.84SS835 pKa = 3.46
MM1 pKa = 7.96SDD3 pKa = 3.1PQEE6 pKa = 3.8RR7 pKa = 11.84SKK9 pKa = 11.34AYY11 pKa = 9.94GLLGEE16 pKa = 4.57RR17 pKa = 11.84LYY19 pKa = 11.36AVASANSHH27 pKa = 5.34MLAGYY32 pKa = 10.42DD33 pKa = 3.81SLDD36 pKa = 3.22FTARR40 pKa = 11.84LVRR43 pKa = 11.84LTGEE47 pKa = 3.93ATALKK52 pKa = 10.39AVDD55 pKa = 4.32PLLPCAVSLLFMDD68 pKa = 6.31FPLQLPCTPEE78 pKa = 3.42EE79 pKa = 3.92TLRR82 pKa = 11.84LVRR85 pKa = 11.84RR86 pKa = 11.84AYY88 pKa = 10.41DD89 pKa = 3.57PNTLEE94 pKa = 4.04EE95 pKa = 4.09VDD97 pKa = 3.89YY98 pKa = 10.23STLSGYY104 pKa = 7.97ATQFARR110 pKa = 11.84VKK112 pKa = 9.46GQRR115 pKa = 11.84GRR117 pKa = 11.84WRR119 pKa = 11.84HH120 pKa = 5.7LGHH123 pKa = 7.31LVCNDD128 pKa = 2.56KK129 pKa = 11.1AFRR132 pKa = 11.84EE133 pKa = 4.38RR134 pKa = 11.84YY135 pKa = 8.97FPKK138 pKa = 10.22KK139 pKa = 9.47KK140 pKa = 9.78HH141 pKa = 5.25AAAAIKK147 pKa = 9.65TNIRR151 pKa = 11.84LGPLARR157 pKa = 11.84AWAARR162 pKa = 11.84YY163 pKa = 9.14GLAALGSHH171 pKa = 7.08LAYY174 pKa = 9.66MVGMPNDD181 pKa = 3.88RR182 pKa = 11.84ACATLLLAQTYY193 pKa = 8.78KK194 pKa = 11.02ARR196 pKa = 11.84FGSEE200 pKa = 3.6GVAWAIASVRR210 pKa = 11.84QPEE213 pKa = 4.2NAKK216 pKa = 10.47GLSNALKK223 pKa = 10.57ALGSNTSEE231 pKa = 4.0PGALFVEE238 pKa = 4.86ANTLQGRR245 pKa = 11.84YY246 pKa = 9.91DD247 pKa = 3.37RR248 pKa = 11.84TLDD251 pKa = 3.26MDD253 pKa = 4.12HH254 pKa = 6.69EE255 pKa = 4.73VEE257 pKa = 4.71SRR259 pKa = 11.84CSPAAIADD267 pKa = 3.62QVIPYY272 pKa = 9.05TDD274 pKa = 3.53EE275 pKa = 4.61LGACIDD281 pKa = 4.91FILDD285 pKa = 3.61TEE287 pKa = 4.73LGGDD291 pKa = 3.88TIEE294 pKa = 6.26LPDD297 pKa = 4.05EE298 pKa = 4.62DD299 pKa = 4.5EE300 pKa = 4.09WWTSRR305 pKa = 11.84WLWCVNGSQNALSDD319 pKa = 3.44KK320 pKa = 11.21ALGIKK325 pKa = 9.88NKK327 pKa = 10.12SGQRR331 pKa = 11.84YY332 pKa = 8.22RR333 pKa = 11.84RR334 pKa = 11.84MAAEE338 pKa = 4.01EE339 pKa = 4.37VNNNPVPAWNGHH351 pKa = 4.63TSVSPSVKK359 pKa = 10.42LEE361 pKa = 3.85NGKK364 pKa = 9.89DD365 pKa = 3.23RR366 pKa = 11.84AIFACDD372 pKa = 3.05TRR374 pKa = 11.84SYY376 pKa = 11.07FAFTYY381 pKa = 9.55WLTPIEE387 pKa = 4.46KK388 pKa = 8.58KK389 pKa = 9.44WRR391 pKa = 11.84GARR394 pKa = 11.84VILNPGEE401 pKa = 4.14GGLYY405 pKa = 8.56GTARR409 pKa = 11.84RR410 pKa = 11.84IRR412 pKa = 11.84GSQTSGGVNLMLDD425 pKa = 3.53YY426 pKa = 11.59DD427 pKa = 4.27NFNSQHH433 pKa = 6.26SNEE436 pKa = 4.15TMAALYY442 pKa = 9.4EE443 pKa = 4.29KK444 pKa = 10.73ALSRR448 pKa = 11.84TNAPAYY454 pKa = 9.99LKK456 pKa = 10.51KK457 pKa = 10.55AVAASVEE464 pKa = 4.3STYY467 pKa = 10.92IHH469 pKa = 6.15YY470 pKa = 10.38KK471 pKa = 9.92GRR473 pKa = 11.84DD474 pKa = 3.08RR475 pKa = 11.84HH476 pKa = 5.76VLGTLMSGHH485 pKa = 7.19RR486 pKa = 11.84ATTFTNSVLNAAYY499 pKa = 8.95ICYY502 pKa = 10.2AVGIPAFKK510 pKa = 10.75RR511 pKa = 11.84MISLHH516 pKa = 6.61AGDD519 pKa = 5.45DD520 pKa = 3.79VYY522 pKa = 11.72LRR524 pKa = 11.84LPTLADD530 pKa = 3.61CATTLNNTKK539 pKa = 10.38RR540 pKa = 11.84VGCRR544 pKa = 11.84MNPTKK549 pKa = 10.55QSIGYY554 pKa = 7.82TGAEE558 pKa = 4.29FLRR561 pKa = 11.84LGINKK566 pKa = 9.61SYY568 pKa = 10.93AIGYY572 pKa = 7.5LCRR575 pKa = 11.84AIASLVSGSWTSLDD589 pKa = 3.4EE590 pKa = 4.23LQPLNALNGAIVQTRR605 pKa = 11.84SCLNRR610 pKa = 11.84GAATGLPEE618 pKa = 5.6LISASFVGLRR628 pKa = 11.84GFKK631 pKa = 10.4RR632 pKa = 11.84RR633 pKa = 11.84DD634 pKa = 3.39LLEE637 pKa = 4.51LLTGVATIKK646 pKa = 10.27PGPVYY651 pKa = 10.29TSSCVIRR658 pKa = 11.84EE659 pKa = 4.17YY660 pKa = 11.04VVEE663 pKa = 4.28QPPPPQFDD671 pKa = 3.89VPPGAGMHH679 pKa = 5.84ATMSYY684 pKa = 10.24LARR687 pKa = 11.84HH688 pKa = 4.9TTLVEE693 pKa = 3.96AQALEE698 pKa = 4.08IARR701 pKa = 11.84PAIKK705 pKa = 10.43SLMLSSSYY713 pKa = 11.11GKK715 pKa = 10.27AGPGAQTRR723 pKa = 11.84PHH725 pKa = 7.54VPMPKK730 pKa = 9.85LRR732 pKa = 11.84RR733 pKa = 11.84QPPRR737 pKa = 11.84VAVGFSMAHH746 pKa = 6.23EE747 pKa = 4.02LTGRR751 pKa = 11.84GVKK754 pKa = 9.67EE755 pKa = 4.3GCLSGHH761 pKa = 6.32PLLRR765 pKa = 11.84LFEE768 pKa = 4.06QRR770 pKa = 11.84LTDD773 pKa = 4.05DD774 pKa = 4.21DD775 pKa = 4.78LRR777 pKa = 11.84ALVALVGGNTSAKK790 pKa = 10.14DD791 pKa = 3.11IRR793 pKa = 11.84AEE795 pKa = 3.89AFGAEE800 pKa = 4.42SSSSTIMGILPHH812 pKa = 6.92SDD814 pKa = 2.3ASNYY818 pKa = 9.62CKK820 pKa = 9.86RR821 pKa = 11.84TRR823 pKa = 11.84NGNIIVPYY831 pKa = 9.8HH832 pKa = 5.86IRR834 pKa = 11.84SS835 pKa = 3.46
Molecular weight: 91.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1607 |
772 |
835 |
803.5 |
86.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.321 ± 0.688 | 1.493 ± 0.308 |
4.729 ± 0.305 | 4.729 ± 0.045 |
3.174 ± 0.393 | 9.272 ± 0.91 |
2.551 ± 0.201 | 3.796 ± 0.114 |
2.427 ± 1.199 | 9.334 ± 1.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.68 ± 0.347 | 4.231 ± 0.058 |
6.845 ± 1.149 | 2.365 ± 0.11 |
7.467 ± 0.057 | 6.472 ± 0.521 |
6.783 ± 0.405 | 6.036 ± 0.472 |
1.307 ± 0.008 | 2.987 ± 0.704 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
