
Human spumaretrovirus (SFVcpz(hu)) (Human foamy virus)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Spumaretrovirinae; Spumavirus; Simian foamy virus
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
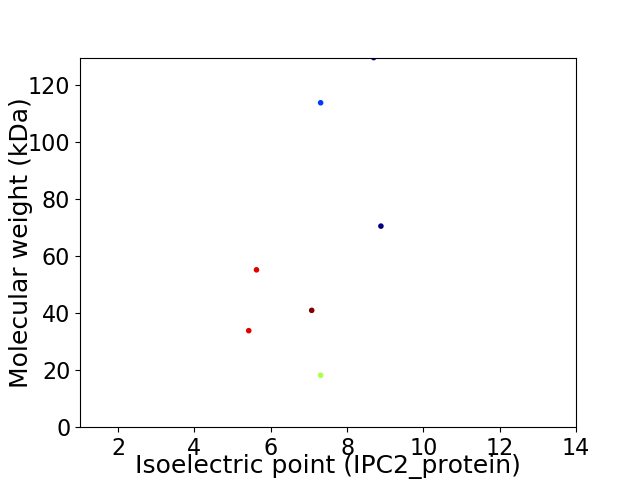
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P14355|BEL3_FOAMV Protein Bel-3 OS=Human spumaretrovirus OX=11963 GN=bel3 PE=1 SV=2
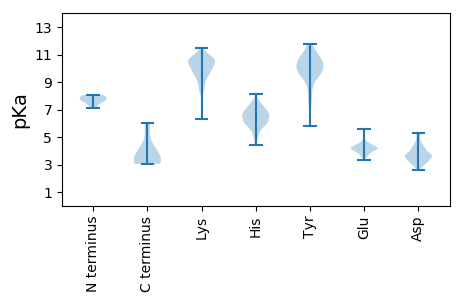
MM1 pKa = 8.05DD2 pKa = 4.83SYY4 pKa = 11.55EE5 pKa = 4.37KK6 pKa = 10.49EE7 pKa = 3.88EE8 pKa = 4.54SVASTSGIQDD18 pKa = 3.7LQTLSEE24 pKa = 4.26LVGPEE29 pKa = 3.64NAGEE33 pKa = 4.39GEE35 pKa = 4.1LTIAEE40 pKa = 4.33EE41 pKa = 4.33PEE43 pKa = 4.04EE44 pKa = 4.15NPRR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84RR51 pKa = 11.84YY52 pKa = 7.28TKK54 pKa = 10.6RR55 pKa = 11.84EE56 pKa = 4.08VKK58 pKa = 10.1CVSYY62 pKa = 10.25HH63 pKa = 6.12AYY65 pKa = 9.92KK66 pKa = 10.4EE67 pKa = 4.3IEE69 pKa = 4.59DD70 pKa = 4.02KK71 pKa = 11.09HH72 pKa = 5.66PQHH75 pKa = 7.08IKK77 pKa = 9.69LQDD80 pKa = 3.76WIPTPEE86 pKa = 4.07EE87 pKa = 3.93MSKK90 pKa = 10.53SLCKK94 pKa = 10.21RR95 pKa = 11.84LILCGLYY102 pKa = 10.29SAEE105 pKa = 4.21KK106 pKa = 9.88ASEE109 pKa = 4.02ILRR112 pKa = 11.84MPFTVSWEE120 pKa = 4.1QSDD123 pKa = 3.85TDD125 pKa = 4.07PDD127 pKa = 4.17CFIVSYY133 pKa = 9.84TCIFCDD139 pKa = 4.74AVIHH143 pKa = 6.87DD144 pKa = 4.8PMPIRR149 pKa = 11.84WDD151 pKa = 3.48PEE153 pKa = 3.45VGIWVKK159 pKa = 10.37YY160 pKa = 9.58KK161 pKa = 9.86PLRR164 pKa = 11.84GIVGSAVFIMHH175 pKa = 6.42KK176 pKa = 9.81HH177 pKa = 4.71QRR179 pKa = 11.84NCSLVKK185 pKa = 10.36PSTSCSEE192 pKa = 4.2GPKK195 pKa = 9.62PRR197 pKa = 11.84PRR199 pKa = 11.84HH200 pKa = 5.92DD201 pKa = 3.25PVLRR205 pKa = 11.84CDD207 pKa = 3.51MFEE210 pKa = 3.94KK211 pKa = 10.14HH212 pKa = 6.28HH213 pKa = 6.71KK214 pKa = 9.43PRR216 pKa = 11.84QKK218 pKa = 9.95RR219 pKa = 11.84PRR221 pKa = 11.84RR222 pKa = 11.84RR223 pKa = 11.84SIDD226 pKa = 3.51NEE228 pKa = 4.28SCASSSDD235 pKa = 3.23TMANEE240 pKa = 4.24PGSLCTNPLWNPGPLLSGLLEE261 pKa = 4.33EE262 pKa = 5.57SSNLPNLEE270 pKa = 3.81VHH272 pKa = 5.92MSGGPFWEE280 pKa = 4.27EE281 pKa = 4.24VYY283 pKa = 10.96GDD285 pKa = 4.66SILGPPSGSGEE296 pKa = 4.05HH297 pKa = 6.54SVLL300 pKa = 4.05
MM1 pKa = 8.05DD2 pKa = 4.83SYY4 pKa = 11.55EE5 pKa = 4.37KK6 pKa = 10.49EE7 pKa = 3.88EE8 pKa = 4.54SVASTSGIQDD18 pKa = 3.7LQTLSEE24 pKa = 4.26LVGPEE29 pKa = 3.64NAGEE33 pKa = 4.39GEE35 pKa = 4.1LTIAEE40 pKa = 4.33EE41 pKa = 4.33PEE43 pKa = 4.04EE44 pKa = 4.15NPRR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84RR51 pKa = 11.84YY52 pKa = 7.28TKK54 pKa = 10.6RR55 pKa = 11.84EE56 pKa = 4.08VKK58 pKa = 10.1CVSYY62 pKa = 10.25HH63 pKa = 6.12AYY65 pKa = 9.92KK66 pKa = 10.4EE67 pKa = 4.3IEE69 pKa = 4.59DD70 pKa = 4.02KK71 pKa = 11.09HH72 pKa = 5.66PQHH75 pKa = 7.08IKK77 pKa = 9.69LQDD80 pKa = 3.76WIPTPEE86 pKa = 4.07EE87 pKa = 3.93MSKK90 pKa = 10.53SLCKK94 pKa = 10.21RR95 pKa = 11.84LILCGLYY102 pKa = 10.29SAEE105 pKa = 4.21KK106 pKa = 9.88ASEE109 pKa = 4.02ILRR112 pKa = 11.84MPFTVSWEE120 pKa = 4.1QSDD123 pKa = 3.85TDD125 pKa = 4.07PDD127 pKa = 4.17CFIVSYY133 pKa = 9.84TCIFCDD139 pKa = 4.74AVIHH143 pKa = 6.87DD144 pKa = 4.8PMPIRR149 pKa = 11.84WDD151 pKa = 3.48PEE153 pKa = 3.45VGIWVKK159 pKa = 10.37YY160 pKa = 9.58KK161 pKa = 9.86PLRR164 pKa = 11.84GIVGSAVFIMHH175 pKa = 6.42KK176 pKa = 9.81HH177 pKa = 4.71QRR179 pKa = 11.84NCSLVKK185 pKa = 10.36PSTSCSEE192 pKa = 4.2GPKK195 pKa = 9.62PRR197 pKa = 11.84PRR199 pKa = 11.84HH200 pKa = 5.92DD201 pKa = 3.25PVLRR205 pKa = 11.84CDD207 pKa = 3.51MFEE210 pKa = 3.94KK211 pKa = 10.14HH212 pKa = 6.28HH213 pKa = 6.71KK214 pKa = 9.43PRR216 pKa = 11.84QKK218 pKa = 9.95RR219 pKa = 11.84PRR221 pKa = 11.84RR222 pKa = 11.84RR223 pKa = 11.84SIDD226 pKa = 3.51NEE228 pKa = 4.28SCASSSDD235 pKa = 3.23TMANEE240 pKa = 4.24PGSLCTNPLWNPGPLLSGLLEE261 pKa = 4.33EE262 pKa = 5.57SSNLPNLEE270 pKa = 3.81VHH272 pKa = 5.92MSGGPFWEE280 pKa = 4.27EE281 pKa = 4.24VYY283 pKa = 10.96GDD285 pKa = 4.66SILGPPSGSGEE296 pKa = 4.05HH297 pKa = 6.54SVLL300 pKa = 4.05
Molecular weight: 33.89 kDa
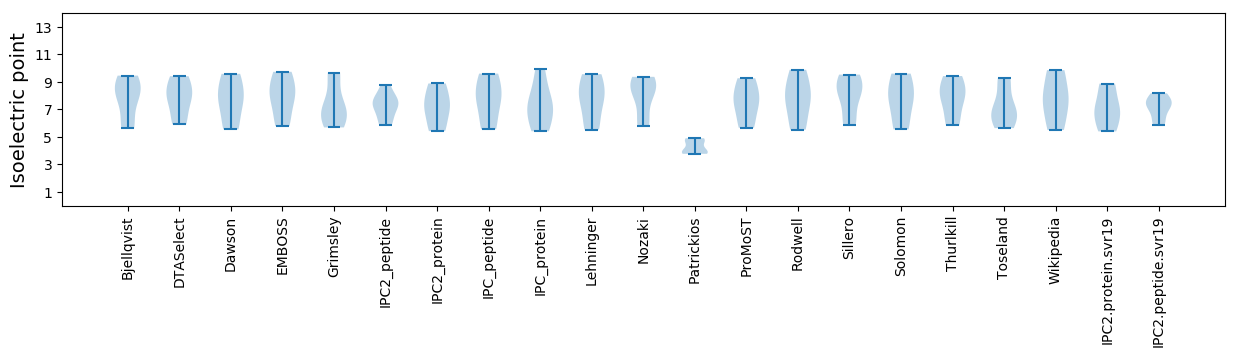
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P14350|POL_FOAMV Pro-Pol polyprotein OS=Human spumaretrovirus OX=11963 GN=pol PE=1 SV=2
MM1 pKa = 7.65ASGSNVEE8 pKa = 4.34EE9 pKa = 4.45YY10 pKa = 10.68EE11 pKa = 4.22LDD13 pKa = 3.64VEE15 pKa = 4.3ALVVILRR22 pKa = 11.84DD23 pKa = 3.56RR24 pKa = 11.84NIPRR28 pKa = 11.84NPLHH32 pKa = 6.68GEE34 pKa = 4.17VIGLRR39 pKa = 11.84LTEE42 pKa = 4.28GWWGQIEE49 pKa = 4.43RR50 pKa = 11.84FQMVRR55 pKa = 11.84LILQDD60 pKa = 4.26DD61 pKa = 4.26DD62 pKa = 5.07NEE64 pKa = 4.13PLQRR68 pKa = 11.84PRR70 pKa = 11.84YY71 pKa = 8.81EE72 pKa = 4.23VIQRR76 pKa = 11.84AVNPHH81 pKa = 4.39TMFMISGPLAEE92 pKa = 5.03LQLAFQDD99 pKa = 3.63LDD101 pKa = 3.93LPEE104 pKa = 5.19GPLRR108 pKa = 11.84FGPLANGHH116 pKa = 5.52YY117 pKa = 10.12VQGDD121 pKa = 4.15PYY123 pKa = 9.92SSSYY127 pKa = 10.97RR128 pKa = 11.84PVTMAEE134 pKa = 4.14TAQMTRR140 pKa = 11.84DD141 pKa = 3.55EE142 pKa = 5.44LEE144 pKa = 4.68DD145 pKa = 3.51VLNTQSEE152 pKa = 4.51IEE154 pKa = 3.87IQMINLLEE162 pKa = 4.42LYY164 pKa = 9.83EE165 pKa = 4.24VEE167 pKa = 3.99TRR169 pKa = 11.84ALRR172 pKa = 11.84RR173 pKa = 11.84QLAEE177 pKa = 4.01RR178 pKa = 11.84SSTGQGGISPGAPRR192 pKa = 11.84SRR194 pKa = 11.84PPVSSFSGLPSLPSIPGIHH213 pKa = 6.77PRR215 pKa = 11.84APSPPRR221 pKa = 11.84ATSTPGNIPWSLGDD235 pKa = 4.42DD236 pKa = 4.06SPPSSSFPGPSQPRR250 pKa = 11.84VSFHH254 pKa = 6.89PGNPFVEE261 pKa = 4.81EE262 pKa = 3.83EE263 pKa = 4.09GHH265 pKa = 6.18RR266 pKa = 11.84PRR268 pKa = 11.84SQSRR272 pKa = 11.84EE273 pKa = 3.56RR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84EE277 pKa = 3.73ILPAPVPSAPPMIQYY292 pKa = 10.08IPVPPPPPIGTVIPIQHH309 pKa = 6.14IRR311 pKa = 11.84SVTGEE316 pKa = 3.86PPRR319 pKa = 11.84NPRR322 pKa = 11.84EE323 pKa = 3.71IPIWLGRR330 pKa = 11.84NAPAIDD336 pKa = 3.66GVFPVTTPDD345 pKa = 3.21LRR347 pKa = 11.84CRR349 pKa = 11.84IINAILGGNIGLSLTPGDD367 pKa = 4.24CLTWDD372 pKa = 3.69SAVATLFIRR381 pKa = 11.84THH383 pKa = 5.47GTFPMHH389 pKa = 6.36QLGNVIKK396 pKa = 10.81GIVDD400 pKa = 3.44QEE402 pKa = 4.65GVATAYY408 pKa = 9.37TLGMMLSGQNYY419 pKa = 8.48QLVSGIIRR427 pKa = 11.84GYY429 pKa = 10.92LPGQAVVTALQQRR442 pKa = 11.84LDD444 pKa = 3.77QEE446 pKa = 4.12IDD448 pKa = 3.17NQTRR452 pKa = 11.84AEE454 pKa = 4.14TFIQHH459 pKa = 6.48LNAVYY464 pKa = 10.31EE465 pKa = 4.1ILGLNARR472 pKa = 11.84GQSIRR477 pKa = 11.84ASVTPQPRR485 pKa = 11.84PSRR488 pKa = 11.84GRR490 pKa = 11.84GRR492 pKa = 11.84GQNTSRR498 pKa = 11.84PSQGPANSGRR508 pKa = 11.84GRR510 pKa = 11.84QRR512 pKa = 11.84PASGQSNRR520 pKa = 11.84GSSTQNQNQDD530 pKa = 3.09NLNQGGYY537 pKa = 7.7NLRR540 pKa = 11.84PRR542 pKa = 11.84TYY544 pKa = 9.41QPQRR548 pKa = 11.84YY549 pKa = 8.66GGGRR553 pKa = 11.84GRR555 pKa = 11.84RR556 pKa = 11.84WNDD559 pKa = 2.73NTNNQEE565 pKa = 4.53SRR567 pKa = 11.84PSDD570 pKa = 3.16QGSQTPRR577 pKa = 11.84PNQAGSGVRR586 pKa = 11.84GNQSQTPRR594 pKa = 11.84PAAGRR599 pKa = 11.84GGRR602 pKa = 11.84GNHH605 pKa = 5.78NRR607 pKa = 11.84NQRR610 pKa = 11.84SSGAGDD616 pKa = 3.14SRR618 pKa = 11.84AVNTVTQSATSSTDD632 pKa = 3.12EE633 pKa = 4.12SSSAVTAASGGDD645 pKa = 3.34QRR647 pKa = 11.84DD648 pKa = 3.15
MM1 pKa = 7.65ASGSNVEE8 pKa = 4.34EE9 pKa = 4.45YY10 pKa = 10.68EE11 pKa = 4.22LDD13 pKa = 3.64VEE15 pKa = 4.3ALVVILRR22 pKa = 11.84DD23 pKa = 3.56RR24 pKa = 11.84NIPRR28 pKa = 11.84NPLHH32 pKa = 6.68GEE34 pKa = 4.17VIGLRR39 pKa = 11.84LTEE42 pKa = 4.28GWWGQIEE49 pKa = 4.43RR50 pKa = 11.84FQMVRR55 pKa = 11.84LILQDD60 pKa = 4.26DD61 pKa = 4.26DD62 pKa = 5.07NEE64 pKa = 4.13PLQRR68 pKa = 11.84PRR70 pKa = 11.84YY71 pKa = 8.81EE72 pKa = 4.23VIQRR76 pKa = 11.84AVNPHH81 pKa = 4.39TMFMISGPLAEE92 pKa = 5.03LQLAFQDD99 pKa = 3.63LDD101 pKa = 3.93LPEE104 pKa = 5.19GPLRR108 pKa = 11.84FGPLANGHH116 pKa = 5.52YY117 pKa = 10.12VQGDD121 pKa = 4.15PYY123 pKa = 9.92SSSYY127 pKa = 10.97RR128 pKa = 11.84PVTMAEE134 pKa = 4.14TAQMTRR140 pKa = 11.84DD141 pKa = 3.55EE142 pKa = 5.44LEE144 pKa = 4.68DD145 pKa = 3.51VLNTQSEE152 pKa = 4.51IEE154 pKa = 3.87IQMINLLEE162 pKa = 4.42LYY164 pKa = 9.83EE165 pKa = 4.24VEE167 pKa = 3.99TRR169 pKa = 11.84ALRR172 pKa = 11.84RR173 pKa = 11.84QLAEE177 pKa = 4.01RR178 pKa = 11.84SSTGQGGISPGAPRR192 pKa = 11.84SRR194 pKa = 11.84PPVSSFSGLPSLPSIPGIHH213 pKa = 6.77PRR215 pKa = 11.84APSPPRR221 pKa = 11.84ATSTPGNIPWSLGDD235 pKa = 4.42DD236 pKa = 4.06SPPSSSFPGPSQPRR250 pKa = 11.84VSFHH254 pKa = 6.89PGNPFVEE261 pKa = 4.81EE262 pKa = 3.83EE263 pKa = 4.09GHH265 pKa = 6.18RR266 pKa = 11.84PRR268 pKa = 11.84SQSRR272 pKa = 11.84EE273 pKa = 3.56RR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84EE277 pKa = 3.73ILPAPVPSAPPMIQYY292 pKa = 10.08IPVPPPPPIGTVIPIQHH309 pKa = 6.14IRR311 pKa = 11.84SVTGEE316 pKa = 3.86PPRR319 pKa = 11.84NPRR322 pKa = 11.84EE323 pKa = 3.71IPIWLGRR330 pKa = 11.84NAPAIDD336 pKa = 3.66GVFPVTTPDD345 pKa = 3.21LRR347 pKa = 11.84CRR349 pKa = 11.84IINAILGGNIGLSLTPGDD367 pKa = 4.24CLTWDD372 pKa = 3.69SAVATLFIRR381 pKa = 11.84THH383 pKa = 5.47GTFPMHH389 pKa = 6.36QLGNVIKK396 pKa = 10.81GIVDD400 pKa = 3.44QEE402 pKa = 4.65GVATAYY408 pKa = 9.37TLGMMLSGQNYY419 pKa = 8.48QLVSGIIRR427 pKa = 11.84GYY429 pKa = 10.92LPGQAVVTALQQRR442 pKa = 11.84LDD444 pKa = 3.77QEE446 pKa = 4.12IDD448 pKa = 3.17NQTRR452 pKa = 11.84AEE454 pKa = 4.14TFIQHH459 pKa = 6.48LNAVYY464 pKa = 10.31EE465 pKa = 4.1ILGLNARR472 pKa = 11.84GQSIRR477 pKa = 11.84ASVTPQPRR485 pKa = 11.84PSRR488 pKa = 11.84GRR490 pKa = 11.84GRR492 pKa = 11.84GQNTSRR498 pKa = 11.84PSQGPANSGRR508 pKa = 11.84GRR510 pKa = 11.84QRR512 pKa = 11.84PASGQSNRR520 pKa = 11.84GSSTQNQNQDD530 pKa = 3.09NLNQGGYY537 pKa = 7.7NLRR540 pKa = 11.84PRR542 pKa = 11.84TYY544 pKa = 9.41QPQRR548 pKa = 11.84YY549 pKa = 8.66GGGRR553 pKa = 11.84GRR555 pKa = 11.84RR556 pKa = 11.84WNDD559 pKa = 2.73NTNNQEE565 pKa = 4.53SRR567 pKa = 11.84PSDD570 pKa = 3.16QGSQTPRR577 pKa = 11.84PNQAGSGVRR586 pKa = 11.84GNQSQTPRR594 pKa = 11.84PAAGRR599 pKa = 11.84GGRR602 pKa = 11.84GNHH605 pKa = 5.78NRR607 pKa = 11.84NQRR610 pKa = 11.84SSGAGDD616 pKa = 3.14SRR618 pKa = 11.84AVNTVTQSATSSTDD632 pKa = 3.12EE633 pKa = 4.12SSSAVTAASGGDD645 pKa = 3.34QRR647 pKa = 11.84DD648 pKa = 3.15
Molecular weight: 70.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4078 |
160 |
1143 |
582.6 |
66.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.321 ± 0.24 | 1.717 ± 0.473 |
4.512 ± 0.33 | 6.106 ± 0.516 |
2.673 ± 0.219 | 6.229 ± 0.963 |
2.55 ± 0.192 | 6.032 ± 0.559 |
5.812 ± 1.11 | 8.95 ± 0.896 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.158 ± 0.294 | 4.586 ± 0.441 |
7.258 ± 0.818 | 5.763 ± 0.415 |
5.468 ± 0.914 | 6.842 ± 0.62 |
6.032 ± 0.361 | 5.836 ± 0.397 |
2.035 ± 0.235 | 4.12 ± 0.537 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
