
Hoeflea marina
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Hoeflea
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
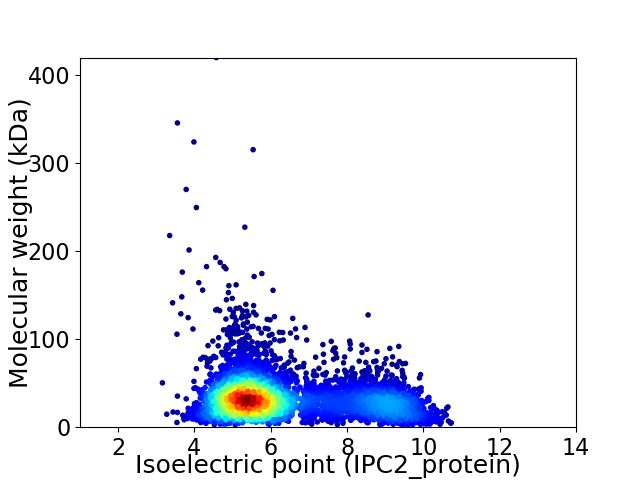
Virtual 2D-PAGE plot for 4798 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
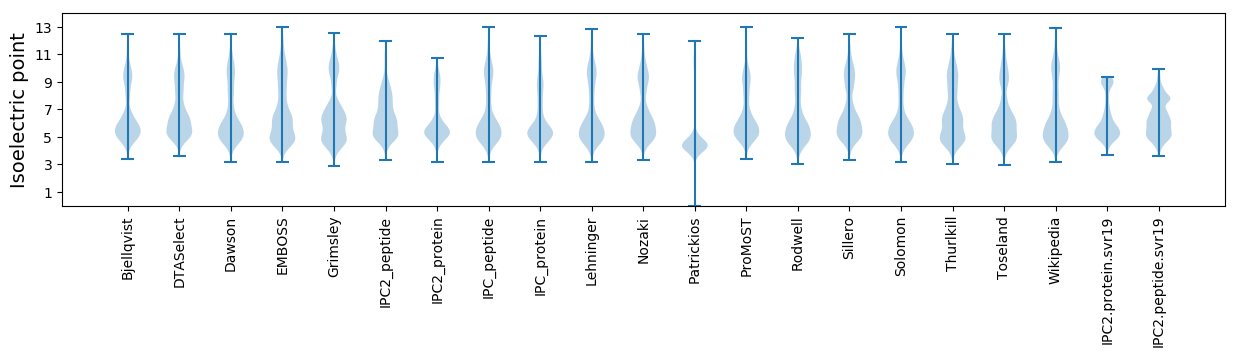
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A317PT36|A0A317PT36_9RHIZ Uncharacterized protein OS=Hoeflea marina OX=274592 GN=DFR52_102649 PE=4 SV=1
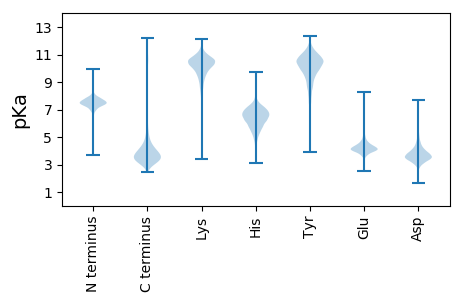
MM1 pKa = 7.74APTLSVRR8 pKa = 11.84VALLRR13 pKa = 11.84HH14 pKa = 6.21GATSSCRR21 pKa = 11.84NAGVEE26 pKa = 3.99RR27 pKa = 11.84HH28 pKa = 5.7RR29 pKa = 11.84SLMRR33 pKa = 11.84LGVAGAALLAGAGAAAADD51 pKa = 3.84DD52 pKa = 4.12VSWINSSTVEE62 pKa = 4.02MRR64 pKa = 11.84WEE66 pKa = 4.74DD67 pKa = 3.35GGNWSGGTVPAQGDD81 pKa = 4.1TVTIATGNSGLYY93 pKa = 9.8TDD95 pKa = 4.07VTVEE99 pKa = 4.14TVNIEE104 pKa = 5.11SGTLWINRR112 pKa = 11.84NAEE115 pKa = 4.11LNADD119 pKa = 3.92TVEE122 pKa = 4.29VTGGRR127 pKa = 11.84LALSQGNPAAPNNSGPQGIANISTLLTISGGTVEE161 pKa = 4.77RR162 pKa = 11.84FGLIEE167 pKa = 4.91LGDD170 pKa = 4.35AGTVIQSGGEE180 pKa = 3.87VLNPVQIHH188 pKa = 5.2TPSYY192 pKa = 8.23VQTGGAMVGLVTASTYY208 pKa = 10.66AISGAEE214 pKa = 4.05GVAATHH220 pKa = 5.88MAGTVTLGTEE230 pKa = 3.95FTMADD235 pKa = 3.39GALVSGTVAGDD246 pKa = 3.64GNATMLQTGGTMDD259 pKa = 4.1GSASGLTSYY268 pKa = 10.93AQTGGTIQGQIGYY281 pKa = 8.01TEE283 pKa = 4.25TFDD286 pKa = 4.58LSGDD290 pKa = 3.77GTVVWGAQLVGDD302 pKa = 4.28SGAVFEE308 pKa = 4.65QSGGTMGGTVQFEE321 pKa = 4.39DD322 pKa = 3.93GAGISKK328 pKa = 9.85YY329 pKa = 9.6VQTGGEE335 pKa = 4.02MTGTVTTGTYY345 pKa = 7.83EE346 pKa = 3.88ASGGTIVVHH355 pKa = 5.97SNVGFSEE362 pKa = 4.36LFALSGDD369 pKa = 3.56ASLGDD374 pKa = 3.64IYY376 pKa = 11.08LIGDD380 pKa = 3.88GDD382 pKa = 4.02AAMTQSGGTMGAWVSGIEE400 pKa = 4.33TYY402 pKa = 11.2TMTGGTILQATEE414 pKa = 4.0VVTDD418 pKa = 4.14AFVQSGGTINDD429 pKa = 3.16ITEE432 pKa = 4.17EE433 pKa = 4.15NDD435 pKa = 2.49WRR437 pKa = 11.84ATVRR441 pKa = 11.84FATEE445 pKa = 3.98YY446 pKa = 10.19EE447 pKa = 4.07LSGDD451 pKa = 3.66ATIGLANLIGGADD464 pKa = 3.29ATMTQSGGTMGGHH477 pKa = 6.25VYY479 pKa = 10.69SEE481 pKa = 4.68TEE483 pKa = 3.73SEE485 pKa = 4.44TIAKK489 pKa = 7.38YY490 pKa = 7.91TQTGGDD496 pKa = 3.22MTGQVTTEE504 pKa = 3.97TYY506 pKa = 8.17EE507 pKa = 4.04ASGGTVSGSVVFSDD521 pKa = 3.98LFAMSGDD528 pKa = 3.45AVVGEE533 pKa = 4.94GYY535 pKa = 10.74SLIGDD540 pKa = 4.22GDD542 pKa = 4.23ALMTQSGGTMAGGVSGIATYY562 pKa = 10.47TMSGGTITGTVDD574 pKa = 3.74FTDD577 pKa = 3.51HH578 pKa = 7.18FEE580 pKa = 4.33LSDD583 pKa = 3.52GRR585 pKa = 11.84IEE587 pKa = 4.15YY588 pKa = 10.7VDD590 pKa = 3.24IVGDD594 pKa = 3.68IDD596 pKa = 3.56ATMTQSGGSMGGAVWSLDD614 pKa = 3.26TDD616 pKa = 4.06DD617 pKa = 4.0YY618 pKa = 11.58TIAKK622 pKa = 7.04YY623 pKa = 7.82TQTGGTMNAEE633 pKa = 4.43VYY635 pKa = 10.72ARR637 pKa = 11.84TYY639 pKa = 10.44DD640 pKa = 3.52VSGGSVDD647 pKa = 3.16GMVRR651 pKa = 11.84FSEE654 pKa = 4.47LFALSGDD661 pKa = 3.67ATVGDD666 pKa = 4.4HH667 pKa = 6.59YY668 pKa = 11.12IWGYY672 pKa = 8.69GAAVATQSGGTMGGVVSDD690 pKa = 3.27ITGYY694 pKa = 6.83TQSGGAMTNYY704 pKa = 9.9VATEE708 pKa = 4.41DD709 pKa = 3.99YY710 pKa = 10.59ALSGGSISGTGHH722 pKa = 6.64VDD724 pKa = 3.67FTDD727 pKa = 4.14LFTLSGSGEE736 pKa = 4.1VLAEE740 pKa = 4.15GTLSGDD746 pKa = 3.35TTAQMAQSGGTMSGHH761 pKa = 5.72VNGITGYY768 pKa = 6.9TQSDD772 pKa = 3.4GTMDD776 pKa = 3.91GKK778 pKa = 11.09VDD780 pKa = 3.43AADD783 pKa = 3.74YY784 pKa = 7.11TQSGGTMGGEE794 pKa = 3.96ATAQTYY800 pKa = 10.9ALAGGTLSGTANFGTSATLSGTGSVTSTGKK830 pKa = 10.97LIGDD834 pKa = 4.38GSATVAQTGGAMDD847 pKa = 3.83GTVTGVTGYY856 pKa = 7.06TQSGGTMGGEE866 pKa = 4.27TTAQTYY872 pKa = 10.39ALSGGTLSGTANFGTSAALSGTGSVTSTGQLIGDD906 pKa = 4.45GSATVAQSGGAMDD919 pKa = 4.04GTVSGVTGYY928 pKa = 7.18TQSGGTMGGKK938 pKa = 8.17VTTGSYY944 pKa = 10.4EE945 pKa = 3.88LLGGVLSGDD954 pKa = 3.66VTFSTLFALSGDD966 pKa = 4.08SVVTSSTQLTQTGGSMDD983 pKa = 3.55STLSGIASYY992 pKa = 8.31TQSGGAMGGDD1002 pKa = 3.74VTATTYY1008 pKa = 9.98TLSGGTLTGKK1018 pKa = 9.99VAGFSSFEE1026 pKa = 4.04LSGDD1030 pKa = 3.54PSDD1033 pKa = 4.87VIVTGTLIGASGSTVTQTGGKK1054 pKa = 8.93MNGNVSGIATYY1065 pKa = 10.09VQQEE1069 pKa = 4.42RR1070 pKa = 11.84VGSAAVISFSEE1081 pKa = 4.84LYY1083 pKa = 11.22DD1084 pKa = 3.6MVDD1087 pKa = 3.55GQVDD1091 pKa = 3.87TGAKK1095 pKa = 9.5IIGGAGSVMTQSGPGKK1111 pKa = 9.63MGGSVTGVASYY1122 pKa = 8.72TQTAGTMNGSVSTGDD1137 pKa = 3.51YY1138 pKa = 9.02TLSAGTVSKK1147 pKa = 9.48TVNFSGAFTLGSIDD1161 pKa = 4.0ANSAGQVNGSAVLTGGAGSTMTQSAGSMKK1190 pKa = 9.64GTVSGVSSYY1199 pKa = 9.6TQTDD1203 pKa = 3.25GTMGGAQDD1211 pKa = 4.09GKK1213 pKa = 10.1PNVIVGKK1220 pKa = 10.41VGTGTYY1226 pKa = 9.73DD1227 pKa = 3.24LSGGTVGGQVNFSTLFALSGTGDD1250 pKa = 3.55VASTATLTGGSGSTMTQSAGTMGGTVTGAATYY1282 pKa = 8.83AQSGGTMSGTVSTGSYY1298 pKa = 9.62EE1299 pKa = 3.99LLGGVLSGDD1308 pKa = 3.66VTFSTLFALSGDD1320 pKa = 3.63GVMTSSTRR1328 pKa = 11.84LTQTGGSMDD1337 pKa = 3.55STLSGIASYY1346 pKa = 8.74TQTGGTMSGATTAASYY1362 pKa = 9.26TLSAGTLSGDD1372 pKa = 3.78ANFSTLFALSGSGDD1386 pKa = 3.39VTAAAEE1392 pKa = 4.03LTGNGEE1398 pKa = 4.3AVMTQSGGTMAGTVTGVTSYY1418 pKa = 8.81TQSGGTMSGATTAASYY1434 pKa = 9.26TLSAGTLSGDD1444 pKa = 3.78ANFSTLFALSGSADD1458 pKa = 3.32VTAAAEE1464 pKa = 4.02LTGNGEE1470 pKa = 4.3AVMTQSGGTMAGTVTGIASYY1490 pKa = 9.79AQSGGTMSGATTAASYY1506 pKa = 9.28TLSAGILSGDD1516 pKa = 3.44ATFSTLLALSGTGGVTSTSQLTGGSGSTMTQSGGTMAGTVTGIASYY1562 pKa = 9.79AQSGGTMSGATTAASYY1578 pKa = 9.26TLSAGTLSGDD1588 pKa = 3.75ANFSTLLALSGTGGVTSTAQLTGGSGSTITQSAGTMAGTVTGIASYY1634 pKa = 8.12TQSGGTMTGATTAASYY1650 pKa = 9.26TLSAGTLSGDD1660 pKa = 3.67ADD1662 pKa = 4.31FSTLFSLSGTGEE1674 pKa = 3.68ISSAAEE1680 pKa = 3.76LTGSGDD1686 pKa = 3.49AAMTQSAGTMNGSVAGAKK1704 pKa = 9.45TYY1706 pKa = 7.49TQSGGTMSGDD1716 pKa = 3.0VAAASFALSGGTLSGDD1732 pKa = 3.2VDD1734 pKa = 4.56FSTLFSLSDD1743 pKa = 3.28TGSIASTALLAGGAGATMTQSGGTMDD1769 pKa = 3.66GTITGIDD1776 pKa = 3.9SYY1778 pKa = 8.91TQSGGTMTGDD1788 pKa = 3.35VTTLAYY1794 pKa = 10.61NHH1796 pKa = 6.4VGGSASGTIDD1806 pKa = 3.05VATYY1810 pKa = 10.37SLSGASASWSGTITASSAFNLGFDD1834 pKa = 4.32GGTATIGAKK1843 pKa = 10.44LSGAGDD1849 pKa = 3.85LVKK1852 pKa = 10.73SGTSTVVLANSANDD1866 pKa = 3.53FSGSVAINGGVLEE1879 pKa = 4.5VRR1881 pKa = 11.84DD1882 pKa = 3.48AALPGEE1888 pKa = 4.38ATVTVADD1895 pKa = 3.91SATLRR1900 pKa = 11.84FNTAAGVSVVFDD1912 pKa = 4.69GIMTGDD1918 pKa = 3.56GGILDD1923 pKa = 3.8KK1924 pKa = 11.2TGLGTLTLGGDD1935 pKa = 3.52IEE1937 pKa = 5.22LGALLVNGGVLNIGNGEE1954 pKa = 3.96PEE1956 pKa = 3.97EE1957 pKa = 5.23ASFQSAYY1964 pKa = 9.62IGVGATVYY1972 pKa = 9.9VASGATLRR1980 pKa = 11.84IRR1982 pKa = 11.84IPQNIANFGTLVNDD1996 pKa = 3.58GTVYY2000 pKa = 11.27DD2001 pKa = 4.84DD2002 pKa = 5.05LDD2004 pKa = 3.7NAGWFEE2010 pKa = 4.28NNALYY2015 pKa = 10.4VANLASNTHH2024 pKa = 7.09DD2025 pKa = 2.74IHH2027 pKa = 7.58NNLRR2031 pKa = 11.84WEE2033 pKa = 4.41GDD2035 pKa = 3.57VLTNNGWIFNNDD2047 pKa = 3.23DD2048 pKa = 3.77ATWVGDD2054 pKa = 3.61IVSNGPLDD2062 pKa = 3.88PNGRR2066 pKa = 11.84PGRR2069 pKa = 11.84IANAGGTWQGDD2080 pKa = 3.47ILSNAGHH2087 pKa = 7.63IINDD2091 pKa = 4.02NIADD2095 pKa = 3.99EE2096 pKa = 4.65SDD2098 pKa = 2.93FGYY2101 pKa = 10.25WFGDD2105 pKa = 3.04IYY2107 pKa = 11.63SNTNWIFNGGGGIWEE2122 pKa = 4.46GDD2124 pKa = 3.86VITNAGRR2131 pKa = 11.84IDD2133 pKa = 3.76NGKK2136 pKa = 9.01SAVWTGDD2143 pKa = 3.33VQSNASTVWNGGIWTGGVLGNAGTLHH2169 pKa = 6.37NAGGTWNGAVEE2180 pKa = 4.53ANTGLIINIAGSDD2193 pKa = 3.61ANGPTGGSTWIGDD2206 pKa = 3.59VTTNAGSILNAADD2219 pKa = 4.02SSWSGNVLANSGTIATLGLWTGDD2242 pKa = 3.21FSNAGRR2248 pKa = 11.84VVAEE2252 pKa = 4.17NEE2254 pKa = 3.98IIGAFSNSGTLRR2266 pKa = 11.84LSGDD2270 pKa = 3.36LSGITALSNSSTIDD2284 pKa = 3.09MKK2286 pKa = 11.31GRR2288 pKa = 11.84GVQVLSVASASFEE2301 pKa = 4.26SGSFLDD2307 pKa = 4.52VGLDD2311 pKa = 3.36SDD2313 pKa = 4.44GNSDD2317 pKa = 3.54QVAVAGLATLGGTVRR2332 pKa = 11.84ISNSGAAHH2340 pKa = 6.6GSGPFTFITAGSFSGSFDD2358 pKa = 4.11AVTTDD2363 pKa = 3.84LAFLAPQLSYY2373 pKa = 11.5GSGSVSVAVQRR2384 pKa = 11.84NDD2386 pKa = 2.77IGFSTTGVTGNQISTGTAVEE2406 pKa = 4.04ALAAGDD2412 pKa = 3.91AVYY2415 pKa = 10.73DD2416 pKa = 3.65AVLWLTEE2423 pKa = 4.38AEE2425 pKa = 4.14AQSAFDD2431 pKa = 3.51QLSGEE2436 pKa = 4.37VHH2438 pKa = 6.84ASHH2441 pKa = 7.16ASAAVEE2447 pKa = 3.99SAAMIGRR2454 pKa = 11.84MASNRR2459 pKa = 11.84IDD2461 pKa = 3.3KK2462 pKa = 10.88AFAAFGEE2469 pKa = 4.81TEE2471 pKa = 4.38ADD2473 pKa = 3.42AQPGSNAVWGRR2484 pKa = 11.84IYY2486 pKa = 10.7GARR2489 pKa = 11.84SLVEE2493 pKa = 4.7AGSQTAGAEE2502 pKa = 4.18TLNGGMVIGLDD2513 pKa = 3.28GMAGVWRR2520 pKa = 11.84LGLLLQGGTSEE2531 pKa = 4.3MEE2533 pKa = 3.6IGALDD2538 pKa = 3.75SSASSDD2544 pKa = 3.47DD2545 pKa = 3.85YY2546 pKa = 12.0GFGAYY2551 pKa = 9.7GGRR2554 pKa = 11.84QFGDD2558 pKa = 3.49TLVSFGAGYY2567 pKa = 8.17TRR2569 pKa = 11.84HH2570 pKa = 6.18DD2571 pKa = 3.59TSTTRR2576 pKa = 11.84QVDD2579 pKa = 3.7FTGFSDD2585 pKa = 3.92TLSADD2590 pKa = 3.83YY2591 pKa = 11.23ASGTAQAFAEE2601 pKa = 4.54VAHH2604 pKa = 6.66RR2605 pKa = 11.84FDD2607 pKa = 4.55LGAVSLTPFAGLAYY2621 pKa = 10.04VRR2623 pKa = 11.84HH2624 pKa = 5.56ATDD2627 pKa = 3.83AFTEE2631 pKa = 4.36TGGAAALSSAANVSEE2646 pKa = 4.12ATFASLGLGVEE2657 pKa = 4.33RR2658 pKa = 11.84QVTVADD2664 pKa = 4.07GMLLTASGSLGWRR2677 pKa = 11.84HH2678 pKa = 6.07AFAEE2682 pKa = 4.67TPISVQRR2689 pKa = 11.84FGSGLAFEE2697 pKa = 5.07ISGEE2701 pKa = 4.05PVASDD2706 pKa = 3.37MLTLGGGLALDD2717 pKa = 3.67IGGGTSLDD2725 pKa = 3.39LTYY2728 pKa = 10.85AGQIGSGAQTHH2739 pKa = 6.87ALQGTWSKK2747 pKa = 11.19RR2748 pKa = 11.84FF2749 pKa = 3.35
MM1 pKa = 7.74APTLSVRR8 pKa = 11.84VALLRR13 pKa = 11.84HH14 pKa = 6.21GATSSCRR21 pKa = 11.84NAGVEE26 pKa = 3.99RR27 pKa = 11.84HH28 pKa = 5.7RR29 pKa = 11.84SLMRR33 pKa = 11.84LGVAGAALLAGAGAAAADD51 pKa = 3.84DD52 pKa = 4.12VSWINSSTVEE62 pKa = 4.02MRR64 pKa = 11.84WEE66 pKa = 4.74DD67 pKa = 3.35GGNWSGGTVPAQGDD81 pKa = 4.1TVTIATGNSGLYY93 pKa = 9.8TDD95 pKa = 4.07VTVEE99 pKa = 4.14TVNIEE104 pKa = 5.11SGTLWINRR112 pKa = 11.84NAEE115 pKa = 4.11LNADD119 pKa = 3.92TVEE122 pKa = 4.29VTGGRR127 pKa = 11.84LALSQGNPAAPNNSGPQGIANISTLLTISGGTVEE161 pKa = 4.77RR162 pKa = 11.84FGLIEE167 pKa = 4.91LGDD170 pKa = 4.35AGTVIQSGGEE180 pKa = 3.87VLNPVQIHH188 pKa = 5.2TPSYY192 pKa = 8.23VQTGGAMVGLVTASTYY208 pKa = 10.66AISGAEE214 pKa = 4.05GVAATHH220 pKa = 5.88MAGTVTLGTEE230 pKa = 3.95FTMADD235 pKa = 3.39GALVSGTVAGDD246 pKa = 3.64GNATMLQTGGTMDD259 pKa = 4.1GSASGLTSYY268 pKa = 10.93AQTGGTIQGQIGYY281 pKa = 8.01TEE283 pKa = 4.25TFDD286 pKa = 4.58LSGDD290 pKa = 3.77GTVVWGAQLVGDD302 pKa = 4.28SGAVFEE308 pKa = 4.65QSGGTMGGTVQFEE321 pKa = 4.39DD322 pKa = 3.93GAGISKK328 pKa = 9.85YY329 pKa = 9.6VQTGGEE335 pKa = 4.02MTGTVTTGTYY345 pKa = 7.83EE346 pKa = 3.88ASGGTIVVHH355 pKa = 5.97SNVGFSEE362 pKa = 4.36LFALSGDD369 pKa = 3.56ASLGDD374 pKa = 3.64IYY376 pKa = 11.08LIGDD380 pKa = 3.88GDD382 pKa = 4.02AAMTQSGGTMGAWVSGIEE400 pKa = 4.33TYY402 pKa = 11.2TMTGGTILQATEE414 pKa = 4.0VVTDD418 pKa = 4.14AFVQSGGTINDD429 pKa = 3.16ITEE432 pKa = 4.17EE433 pKa = 4.15NDD435 pKa = 2.49WRR437 pKa = 11.84ATVRR441 pKa = 11.84FATEE445 pKa = 3.98YY446 pKa = 10.19EE447 pKa = 4.07LSGDD451 pKa = 3.66ATIGLANLIGGADD464 pKa = 3.29ATMTQSGGTMGGHH477 pKa = 6.25VYY479 pKa = 10.69SEE481 pKa = 4.68TEE483 pKa = 3.73SEE485 pKa = 4.44TIAKK489 pKa = 7.38YY490 pKa = 7.91TQTGGDD496 pKa = 3.22MTGQVTTEE504 pKa = 3.97TYY506 pKa = 8.17EE507 pKa = 4.04ASGGTVSGSVVFSDD521 pKa = 3.98LFAMSGDD528 pKa = 3.45AVVGEE533 pKa = 4.94GYY535 pKa = 10.74SLIGDD540 pKa = 4.22GDD542 pKa = 4.23ALMTQSGGTMAGGVSGIATYY562 pKa = 10.47TMSGGTITGTVDD574 pKa = 3.74FTDD577 pKa = 3.51HH578 pKa = 7.18FEE580 pKa = 4.33LSDD583 pKa = 3.52GRR585 pKa = 11.84IEE587 pKa = 4.15YY588 pKa = 10.7VDD590 pKa = 3.24IVGDD594 pKa = 3.68IDD596 pKa = 3.56ATMTQSGGSMGGAVWSLDD614 pKa = 3.26TDD616 pKa = 4.06DD617 pKa = 4.0YY618 pKa = 11.58TIAKK622 pKa = 7.04YY623 pKa = 7.82TQTGGTMNAEE633 pKa = 4.43VYY635 pKa = 10.72ARR637 pKa = 11.84TYY639 pKa = 10.44DD640 pKa = 3.52VSGGSVDD647 pKa = 3.16GMVRR651 pKa = 11.84FSEE654 pKa = 4.47LFALSGDD661 pKa = 3.67ATVGDD666 pKa = 4.4HH667 pKa = 6.59YY668 pKa = 11.12IWGYY672 pKa = 8.69GAAVATQSGGTMGGVVSDD690 pKa = 3.27ITGYY694 pKa = 6.83TQSGGAMTNYY704 pKa = 9.9VATEE708 pKa = 4.41DD709 pKa = 3.99YY710 pKa = 10.59ALSGGSISGTGHH722 pKa = 6.64VDD724 pKa = 3.67FTDD727 pKa = 4.14LFTLSGSGEE736 pKa = 4.1VLAEE740 pKa = 4.15GTLSGDD746 pKa = 3.35TTAQMAQSGGTMSGHH761 pKa = 5.72VNGITGYY768 pKa = 6.9TQSDD772 pKa = 3.4GTMDD776 pKa = 3.91GKK778 pKa = 11.09VDD780 pKa = 3.43AADD783 pKa = 3.74YY784 pKa = 7.11TQSGGTMGGEE794 pKa = 3.96ATAQTYY800 pKa = 10.9ALAGGTLSGTANFGTSATLSGTGSVTSTGKK830 pKa = 10.97LIGDD834 pKa = 4.38GSATVAQTGGAMDD847 pKa = 3.83GTVTGVTGYY856 pKa = 7.06TQSGGTMGGEE866 pKa = 4.27TTAQTYY872 pKa = 10.39ALSGGTLSGTANFGTSAALSGTGSVTSTGQLIGDD906 pKa = 4.45GSATVAQSGGAMDD919 pKa = 4.04GTVSGVTGYY928 pKa = 7.18TQSGGTMGGKK938 pKa = 8.17VTTGSYY944 pKa = 10.4EE945 pKa = 3.88LLGGVLSGDD954 pKa = 3.66VTFSTLFALSGDD966 pKa = 4.08SVVTSSTQLTQTGGSMDD983 pKa = 3.55STLSGIASYY992 pKa = 8.31TQSGGAMGGDD1002 pKa = 3.74VTATTYY1008 pKa = 9.98TLSGGTLTGKK1018 pKa = 9.99VAGFSSFEE1026 pKa = 4.04LSGDD1030 pKa = 3.54PSDD1033 pKa = 4.87VIVTGTLIGASGSTVTQTGGKK1054 pKa = 8.93MNGNVSGIATYY1065 pKa = 10.09VQQEE1069 pKa = 4.42RR1070 pKa = 11.84VGSAAVISFSEE1081 pKa = 4.84LYY1083 pKa = 11.22DD1084 pKa = 3.6MVDD1087 pKa = 3.55GQVDD1091 pKa = 3.87TGAKK1095 pKa = 9.5IIGGAGSVMTQSGPGKK1111 pKa = 9.63MGGSVTGVASYY1122 pKa = 8.72TQTAGTMNGSVSTGDD1137 pKa = 3.51YY1138 pKa = 9.02TLSAGTVSKK1147 pKa = 9.48TVNFSGAFTLGSIDD1161 pKa = 4.0ANSAGQVNGSAVLTGGAGSTMTQSAGSMKK1190 pKa = 9.64GTVSGVSSYY1199 pKa = 9.6TQTDD1203 pKa = 3.25GTMGGAQDD1211 pKa = 4.09GKK1213 pKa = 10.1PNVIVGKK1220 pKa = 10.41VGTGTYY1226 pKa = 9.73DD1227 pKa = 3.24LSGGTVGGQVNFSTLFALSGTGDD1250 pKa = 3.55VASTATLTGGSGSTMTQSAGTMGGTVTGAATYY1282 pKa = 8.83AQSGGTMSGTVSTGSYY1298 pKa = 9.62EE1299 pKa = 3.99LLGGVLSGDD1308 pKa = 3.66VTFSTLFALSGDD1320 pKa = 3.63GVMTSSTRR1328 pKa = 11.84LTQTGGSMDD1337 pKa = 3.55STLSGIASYY1346 pKa = 8.74TQTGGTMSGATTAASYY1362 pKa = 9.26TLSAGTLSGDD1372 pKa = 3.78ANFSTLFALSGSGDD1386 pKa = 3.39VTAAAEE1392 pKa = 4.03LTGNGEE1398 pKa = 4.3AVMTQSGGTMAGTVTGVTSYY1418 pKa = 8.81TQSGGTMSGATTAASYY1434 pKa = 9.26TLSAGTLSGDD1444 pKa = 3.78ANFSTLFALSGSADD1458 pKa = 3.32VTAAAEE1464 pKa = 4.02LTGNGEE1470 pKa = 4.3AVMTQSGGTMAGTVTGIASYY1490 pKa = 9.79AQSGGTMSGATTAASYY1506 pKa = 9.28TLSAGILSGDD1516 pKa = 3.44ATFSTLLALSGTGGVTSTSQLTGGSGSTMTQSGGTMAGTVTGIASYY1562 pKa = 9.79AQSGGTMSGATTAASYY1578 pKa = 9.26TLSAGTLSGDD1588 pKa = 3.75ANFSTLLALSGTGGVTSTAQLTGGSGSTITQSAGTMAGTVTGIASYY1634 pKa = 8.12TQSGGTMTGATTAASYY1650 pKa = 9.26TLSAGTLSGDD1660 pKa = 3.67ADD1662 pKa = 4.31FSTLFSLSGTGEE1674 pKa = 3.68ISSAAEE1680 pKa = 3.76LTGSGDD1686 pKa = 3.49AAMTQSAGTMNGSVAGAKK1704 pKa = 9.45TYY1706 pKa = 7.49TQSGGTMSGDD1716 pKa = 3.0VAAASFALSGGTLSGDD1732 pKa = 3.2VDD1734 pKa = 4.56FSTLFSLSDD1743 pKa = 3.28TGSIASTALLAGGAGATMTQSGGTMDD1769 pKa = 3.66GTITGIDD1776 pKa = 3.9SYY1778 pKa = 8.91TQSGGTMTGDD1788 pKa = 3.35VTTLAYY1794 pKa = 10.61NHH1796 pKa = 6.4VGGSASGTIDD1806 pKa = 3.05VATYY1810 pKa = 10.37SLSGASASWSGTITASSAFNLGFDD1834 pKa = 4.32GGTATIGAKK1843 pKa = 10.44LSGAGDD1849 pKa = 3.85LVKK1852 pKa = 10.73SGTSTVVLANSANDD1866 pKa = 3.53FSGSVAINGGVLEE1879 pKa = 4.5VRR1881 pKa = 11.84DD1882 pKa = 3.48AALPGEE1888 pKa = 4.38ATVTVADD1895 pKa = 3.91SATLRR1900 pKa = 11.84FNTAAGVSVVFDD1912 pKa = 4.69GIMTGDD1918 pKa = 3.56GGILDD1923 pKa = 3.8KK1924 pKa = 11.2TGLGTLTLGGDD1935 pKa = 3.52IEE1937 pKa = 5.22LGALLVNGGVLNIGNGEE1954 pKa = 3.96PEE1956 pKa = 3.97EE1957 pKa = 5.23ASFQSAYY1964 pKa = 9.62IGVGATVYY1972 pKa = 9.9VASGATLRR1980 pKa = 11.84IRR1982 pKa = 11.84IPQNIANFGTLVNDD1996 pKa = 3.58GTVYY2000 pKa = 11.27DD2001 pKa = 4.84DD2002 pKa = 5.05LDD2004 pKa = 3.7NAGWFEE2010 pKa = 4.28NNALYY2015 pKa = 10.4VANLASNTHH2024 pKa = 7.09DD2025 pKa = 2.74IHH2027 pKa = 7.58NNLRR2031 pKa = 11.84WEE2033 pKa = 4.41GDD2035 pKa = 3.57VLTNNGWIFNNDD2047 pKa = 3.23DD2048 pKa = 3.77ATWVGDD2054 pKa = 3.61IVSNGPLDD2062 pKa = 3.88PNGRR2066 pKa = 11.84PGRR2069 pKa = 11.84IANAGGTWQGDD2080 pKa = 3.47ILSNAGHH2087 pKa = 7.63IINDD2091 pKa = 4.02NIADD2095 pKa = 3.99EE2096 pKa = 4.65SDD2098 pKa = 2.93FGYY2101 pKa = 10.25WFGDD2105 pKa = 3.04IYY2107 pKa = 11.63SNTNWIFNGGGGIWEE2122 pKa = 4.46GDD2124 pKa = 3.86VITNAGRR2131 pKa = 11.84IDD2133 pKa = 3.76NGKK2136 pKa = 9.01SAVWTGDD2143 pKa = 3.33VQSNASTVWNGGIWTGGVLGNAGTLHH2169 pKa = 6.37NAGGTWNGAVEE2180 pKa = 4.53ANTGLIINIAGSDD2193 pKa = 3.61ANGPTGGSTWIGDD2206 pKa = 3.59VTTNAGSILNAADD2219 pKa = 4.02SSWSGNVLANSGTIATLGLWTGDD2242 pKa = 3.21FSNAGRR2248 pKa = 11.84VVAEE2252 pKa = 4.17NEE2254 pKa = 3.98IIGAFSNSGTLRR2266 pKa = 11.84LSGDD2270 pKa = 3.36LSGITALSNSSTIDD2284 pKa = 3.09MKK2286 pKa = 11.31GRR2288 pKa = 11.84GVQVLSVASASFEE2301 pKa = 4.26SGSFLDD2307 pKa = 4.52VGLDD2311 pKa = 3.36SDD2313 pKa = 4.44GNSDD2317 pKa = 3.54QVAVAGLATLGGTVRR2332 pKa = 11.84ISNSGAAHH2340 pKa = 6.6GSGPFTFITAGSFSGSFDD2358 pKa = 4.11AVTTDD2363 pKa = 3.84LAFLAPQLSYY2373 pKa = 11.5GSGSVSVAVQRR2384 pKa = 11.84NDD2386 pKa = 2.77IGFSTTGVTGNQISTGTAVEE2406 pKa = 4.04ALAAGDD2412 pKa = 3.91AVYY2415 pKa = 10.73DD2416 pKa = 3.65AVLWLTEE2423 pKa = 4.38AEE2425 pKa = 4.14AQSAFDD2431 pKa = 3.51QLSGEE2436 pKa = 4.37VHH2438 pKa = 6.84ASHH2441 pKa = 7.16ASAAVEE2447 pKa = 3.99SAAMIGRR2454 pKa = 11.84MASNRR2459 pKa = 11.84IDD2461 pKa = 3.3KK2462 pKa = 10.88AFAAFGEE2469 pKa = 4.81TEE2471 pKa = 4.38ADD2473 pKa = 3.42AQPGSNAVWGRR2484 pKa = 11.84IYY2486 pKa = 10.7GARR2489 pKa = 11.84SLVEE2493 pKa = 4.7AGSQTAGAEE2502 pKa = 4.18TLNGGMVIGLDD2513 pKa = 3.28GMAGVWRR2520 pKa = 11.84LGLLLQGGTSEE2531 pKa = 4.3MEE2533 pKa = 3.6IGALDD2538 pKa = 3.75SSASSDD2544 pKa = 3.47DD2545 pKa = 3.85YY2546 pKa = 12.0GFGAYY2551 pKa = 9.7GGRR2554 pKa = 11.84QFGDD2558 pKa = 3.49TLVSFGAGYY2567 pKa = 8.17TRR2569 pKa = 11.84HH2570 pKa = 6.18DD2571 pKa = 3.59TSTTRR2576 pKa = 11.84QVDD2579 pKa = 3.7FTGFSDD2585 pKa = 3.92TLSADD2590 pKa = 3.83YY2591 pKa = 11.23ASGTAQAFAEE2601 pKa = 4.54VAHH2604 pKa = 6.66RR2605 pKa = 11.84FDD2607 pKa = 4.55LGAVSLTPFAGLAYY2621 pKa = 10.04VRR2623 pKa = 11.84HH2624 pKa = 5.56ATDD2627 pKa = 3.83AFTEE2631 pKa = 4.36TGGAAALSSAANVSEE2646 pKa = 4.12ATFASLGLGVEE2657 pKa = 4.33RR2658 pKa = 11.84QVTVADD2664 pKa = 4.07GMLLTASGSLGWRR2677 pKa = 11.84HH2678 pKa = 6.07AFAEE2682 pKa = 4.67TPISVQRR2689 pKa = 11.84FGSGLAFEE2697 pKa = 5.07ISGEE2701 pKa = 4.05PVASDD2706 pKa = 3.37MLTLGGGLALDD2717 pKa = 3.67IGGGTSLDD2725 pKa = 3.39LTYY2728 pKa = 10.85AGQIGSGAQTHH2739 pKa = 6.87ALQGTWSKK2747 pKa = 11.19RR2748 pKa = 11.84FF2749 pKa = 3.35
Molecular weight: 270.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A317PV17|A0A317PV17_9RHIZ DHA1 family inner membrane transport protein OS=Hoeflea marina OX=274592 GN=DFR52_1011233 PE=4 SV=1
MM1 pKa = 7.42TNQATGLSIATVAALIGDD19 pKa = 4.14VARR22 pKa = 11.84SNILHH27 pKa = 6.84ALMGGKK33 pKa = 9.92ALTATEE39 pKa = 4.4LAHH42 pKa = 6.28HH43 pKa = 7.02AGVSAPTTSGHH54 pKa = 6.72LARR57 pKa = 11.84LVDD60 pKa = 3.93GKK62 pKa = 10.89LVLMEE67 pKa = 3.68KK68 pKa = 10.01QGRR71 pKa = 11.84HH72 pKa = 4.43RR73 pKa = 11.84YY74 pKa = 8.7FRR76 pKa = 11.84LASPAVAEE84 pKa = 4.07ILEE87 pKa = 4.33GLGNLAAAGPARR99 pKa = 11.84HH100 pKa = 6.17RR101 pKa = 11.84PTGPKK106 pKa = 9.86DD107 pKa = 2.97LAMRR111 pKa = 11.84HH112 pKa = 5.84ARR114 pKa = 11.84TCYY117 pKa = 10.02DD118 pKa = 2.98HH119 pKa = 6.17MAGRR123 pKa = 11.84IAVALTDD130 pKa = 3.68SLTEE134 pKa = 3.82RR135 pKa = 11.84GLIIIEE141 pKa = 4.33DD142 pKa = 3.71RR143 pKa = 11.84SGLVTDD149 pKa = 4.19DD150 pKa = 3.16GRR152 pKa = 11.84RR153 pKa = 11.84FFAGFGIDD161 pKa = 4.61LEE163 pKa = 4.75SKK165 pKa = 10.26PRR167 pKa = 11.84TARR170 pKa = 11.84PLCRR174 pKa = 11.84TCLDD178 pKa = 2.96WSEE181 pKa = 4.48RR182 pKa = 11.84RR183 pKa = 11.84MHH185 pKa = 6.83LGGRR189 pKa = 11.84LGASLLARR197 pKa = 11.84SIEE200 pKa = 4.24RR201 pKa = 11.84GWVRR205 pKa = 11.84TIRR208 pKa = 11.84DD209 pKa = 3.26SRR211 pKa = 11.84ALEE214 pKa = 3.79VTRR217 pKa = 11.84AGEE220 pKa = 3.94RR221 pKa = 11.84GFTEE225 pKa = 4.08TFGIASEE232 pKa = 4.09MLRR235 pKa = 11.84PPRR238 pKa = 4.55
MM1 pKa = 7.42TNQATGLSIATVAALIGDD19 pKa = 4.14VARR22 pKa = 11.84SNILHH27 pKa = 6.84ALMGGKK33 pKa = 9.92ALTATEE39 pKa = 4.4LAHH42 pKa = 6.28HH43 pKa = 7.02AGVSAPTTSGHH54 pKa = 6.72LARR57 pKa = 11.84LVDD60 pKa = 3.93GKK62 pKa = 10.89LVLMEE67 pKa = 3.68KK68 pKa = 10.01QGRR71 pKa = 11.84HH72 pKa = 4.43RR73 pKa = 11.84YY74 pKa = 8.7FRR76 pKa = 11.84LASPAVAEE84 pKa = 4.07ILEE87 pKa = 4.33GLGNLAAAGPARR99 pKa = 11.84HH100 pKa = 6.17RR101 pKa = 11.84PTGPKK106 pKa = 9.86DD107 pKa = 2.97LAMRR111 pKa = 11.84HH112 pKa = 5.84ARR114 pKa = 11.84TCYY117 pKa = 10.02DD118 pKa = 2.98HH119 pKa = 6.17MAGRR123 pKa = 11.84IAVALTDD130 pKa = 3.68SLTEE134 pKa = 3.82RR135 pKa = 11.84GLIIIEE141 pKa = 4.33DD142 pKa = 3.71RR143 pKa = 11.84SGLVTDD149 pKa = 4.19DD150 pKa = 3.16GRR152 pKa = 11.84RR153 pKa = 11.84FFAGFGIDD161 pKa = 4.61LEE163 pKa = 4.75SKK165 pKa = 10.26PRR167 pKa = 11.84TARR170 pKa = 11.84PLCRR174 pKa = 11.84TCLDD178 pKa = 2.96WSEE181 pKa = 4.48RR182 pKa = 11.84RR183 pKa = 11.84MHH185 pKa = 6.83LGGRR189 pKa = 11.84LGASLLARR197 pKa = 11.84SIEE200 pKa = 4.24RR201 pKa = 11.84GWVRR205 pKa = 11.84TIRR208 pKa = 11.84DD209 pKa = 3.26SRR211 pKa = 11.84ALEE214 pKa = 3.79VTRR217 pKa = 11.84AGEE220 pKa = 3.94RR221 pKa = 11.84GFTEE225 pKa = 4.08TFGIASEE232 pKa = 4.09MLRR235 pKa = 11.84PPRR238 pKa = 4.55
Molecular weight: 25.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1545255 |
27 |
3958 |
322.1 |
34.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.586 ± 0.044 | 0.828 ± 0.013 |
5.882 ± 0.034 | 5.447 ± 0.034 |
3.813 ± 0.025 | 8.923 ± 0.054 |
2.031 ± 0.021 | 5.514 ± 0.029 |
2.914 ± 0.027 | 9.945 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.707 ± 0.019 | 2.587 ± 0.023 |
4.983 ± 0.031 | 2.863 ± 0.018 |
6.954 ± 0.048 | 5.866 ± 0.033 |
5.349 ± 0.042 | 7.369 ± 0.029 |
1.276 ± 0.014 | 2.165 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
