
Candidatus Termititenax aidoneus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Candidatus Margulisbacteria; Candidatus Termititenacia; Candidatus Termititenacales; Candidatus Termititenacaceae; Candidatus Termititenax
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
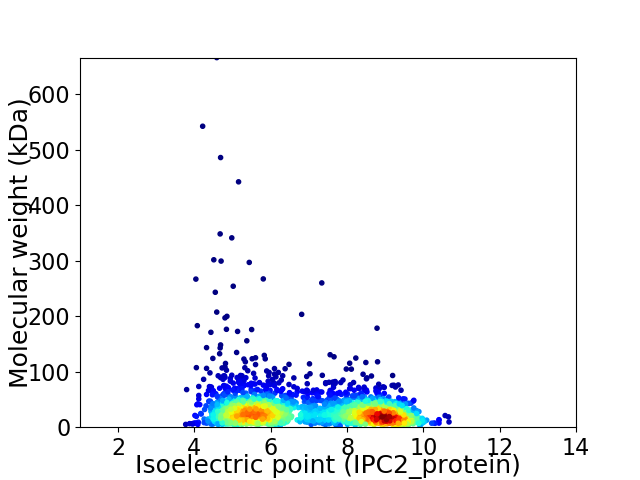
Virtual 2D-PAGE plot for 2306 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A388TG92|A0A388TG92_9BACT Uncharacterized protein OS=Candidatus Termititenax aidoneus OX=2218524 GN=NO1_2225 PE=4 SV=1
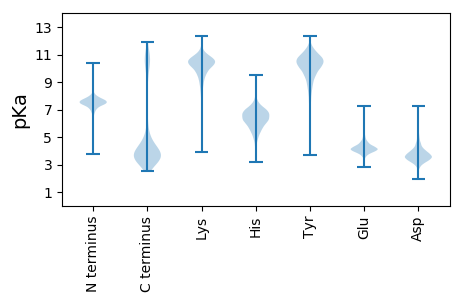
MM1 pKa = 7.89LDD3 pKa = 3.05NYY5 pKa = 10.67EE6 pKa = 4.38YY7 pKa = 10.55IDD9 pKa = 4.37LEE11 pKa = 4.36FVYY14 pKa = 10.23TDD16 pKa = 3.43PQTGVLRR23 pKa = 11.84NLGGITDD30 pKa = 3.86NQDD33 pKa = 3.38LLFFEE38 pKa = 5.05SVAVAKK44 pKa = 10.63
MM1 pKa = 7.89LDD3 pKa = 3.05NYY5 pKa = 10.67EE6 pKa = 4.38YY7 pKa = 10.55IDD9 pKa = 4.37LEE11 pKa = 4.36FVYY14 pKa = 10.23TDD16 pKa = 3.43PQTGVLRR23 pKa = 11.84NLGGITDD30 pKa = 3.86NQDD33 pKa = 3.38LLFFEE38 pKa = 5.05SVAVAKK44 pKa = 10.63
Molecular weight: 5.03 kDa
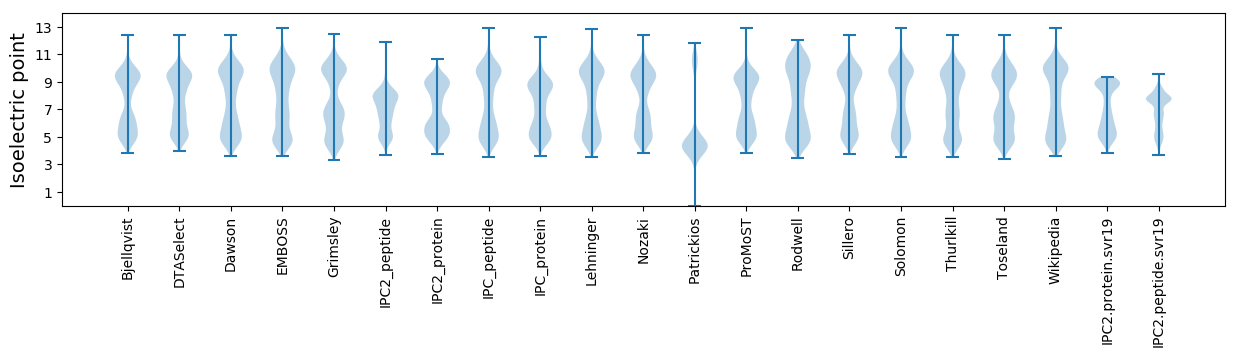
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A388T7M3|A0A388T7M3_9BACT ATP synthase subunit alpha OS=Candidatus Termititenax aidoneus OX=2218524 GN=atpA PE=3 SV=1
MM1 pKa = 7.7KK2 pKa = 10.13ILLAGYY8 pKa = 10.07YY9 pKa = 10.75GFGNLGDD16 pKa = 4.07EE17 pKa = 5.01LLAQTARR24 pKa = 11.84EE25 pKa = 4.05LLSAKK30 pKa = 9.05HH31 pKa = 5.21TVTVCRR37 pKa = 11.84ARR39 pKa = 11.84RR40 pKa = 11.84NFLFLLLPADD50 pKa = 4.35CLLLAGGSLFQDD62 pKa = 3.28VTGRR66 pKa = 11.84GLSVLYY72 pKa = 9.98YY73 pKa = 10.3SAWACAAKK81 pKa = 10.56LFRR84 pKa = 11.84KK85 pKa = 9.94KK86 pKa = 10.68LFLVAQGIGPIRR98 pKa = 11.84APLNRR103 pKa = 11.84WLTRR107 pKa = 11.84AVFRR111 pKa = 11.84RR112 pKa = 11.84ADD114 pKa = 3.77FVSLRR119 pKa = 11.84DD120 pKa = 3.58RR121 pKa = 11.84EE122 pKa = 4.39SAEE125 pKa = 4.05FLGLPQSHH133 pKa = 6.25LTADD137 pKa = 3.97LLFAAKK143 pKa = 10.22LRR145 pKa = 11.84TLKK148 pKa = 9.57TAQRR152 pKa = 11.84QQAKK156 pKa = 7.61TAPLPVGAARR166 pKa = 11.84RR167 pKa = 11.84RR168 pKa = 11.84QARR171 pKa = 11.84AARR174 pKa = 11.84RR175 pKa = 11.84RR176 pKa = 11.84QIIVNLKK183 pKa = 10.04KK184 pKa = 10.42SAPQILTEE192 pKa = 4.02FQPVYY197 pKa = 10.94LAMQPSADD205 pKa = 3.58WGRR208 pKa = 11.84PCAPPDD214 pKa = 3.44FAAARR219 pKa = 11.84LAVGMRR225 pKa = 11.84LHH227 pKa = 6.44FLILAVLHH235 pKa = 5.97NVPAVGIAYY244 pKa = 9.38DD245 pKa = 3.58PKK247 pKa = 11.03VEE249 pKa = 4.07NFCRR253 pKa = 11.84KK254 pKa = 9.24FDD256 pKa = 3.97LPYY259 pKa = 9.59VTLDD263 pKa = 3.19EE264 pKa = 5.16LEE266 pKa = 4.76RR267 pKa = 11.84LPQIIRR273 pKa = 11.84RR274 pKa = 11.84EE275 pKa = 3.99LAKK278 pKa = 10.2PAAAKK283 pKa = 10.09KK284 pKa = 10.13RR285 pKa = 11.84LAVLVKK291 pKa = 8.44QEE293 pKa = 3.58RR294 pKa = 11.84RR295 pKa = 11.84LARR298 pKa = 11.84EE299 pKa = 3.94GVKK302 pKa = 10.57VLLEE306 pKa = 4.05KK307 pKa = 11.25LNGG310 pKa = 3.58
MM1 pKa = 7.7KK2 pKa = 10.13ILLAGYY8 pKa = 10.07YY9 pKa = 10.75GFGNLGDD16 pKa = 4.07EE17 pKa = 5.01LLAQTARR24 pKa = 11.84EE25 pKa = 4.05LLSAKK30 pKa = 9.05HH31 pKa = 5.21TVTVCRR37 pKa = 11.84ARR39 pKa = 11.84RR40 pKa = 11.84NFLFLLLPADD50 pKa = 4.35CLLLAGGSLFQDD62 pKa = 3.28VTGRR66 pKa = 11.84GLSVLYY72 pKa = 9.98YY73 pKa = 10.3SAWACAAKK81 pKa = 10.56LFRR84 pKa = 11.84KK85 pKa = 9.94KK86 pKa = 10.68LFLVAQGIGPIRR98 pKa = 11.84APLNRR103 pKa = 11.84WLTRR107 pKa = 11.84AVFRR111 pKa = 11.84RR112 pKa = 11.84ADD114 pKa = 3.77FVSLRR119 pKa = 11.84DD120 pKa = 3.58RR121 pKa = 11.84EE122 pKa = 4.39SAEE125 pKa = 4.05FLGLPQSHH133 pKa = 6.25LTADD137 pKa = 3.97LLFAAKK143 pKa = 10.22LRR145 pKa = 11.84TLKK148 pKa = 9.57TAQRR152 pKa = 11.84QQAKK156 pKa = 7.61TAPLPVGAARR166 pKa = 11.84RR167 pKa = 11.84RR168 pKa = 11.84QARR171 pKa = 11.84AARR174 pKa = 11.84RR175 pKa = 11.84RR176 pKa = 11.84QIIVNLKK183 pKa = 10.04KK184 pKa = 10.42SAPQILTEE192 pKa = 4.02FQPVYY197 pKa = 10.94LAMQPSADD205 pKa = 3.58WGRR208 pKa = 11.84PCAPPDD214 pKa = 3.44FAAARR219 pKa = 11.84LAVGMRR225 pKa = 11.84LHH227 pKa = 6.44FLILAVLHH235 pKa = 5.97NVPAVGIAYY244 pKa = 9.38DD245 pKa = 3.58PKK247 pKa = 11.03VEE249 pKa = 4.07NFCRR253 pKa = 11.84KK254 pKa = 9.24FDD256 pKa = 3.97LPYY259 pKa = 9.59VTLDD263 pKa = 3.19EE264 pKa = 5.16LEE266 pKa = 4.76RR267 pKa = 11.84LPQIIRR273 pKa = 11.84RR274 pKa = 11.84EE275 pKa = 3.99LAKK278 pKa = 10.2PAAAKK283 pKa = 10.09KK284 pKa = 10.13RR285 pKa = 11.84LAVLVKK291 pKa = 8.44QEE293 pKa = 3.58RR294 pKa = 11.84RR295 pKa = 11.84LARR298 pKa = 11.84EE299 pKa = 3.94GVKK302 pKa = 10.57VLLEE306 pKa = 4.05KK307 pKa = 11.25LNGG310 pKa = 3.58
Molecular weight: 34.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
677573 |
20 |
6012 |
293.8 |
32.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.402 ± 0.07 | 0.957 ± 0.025 |
5.407 ± 0.046 | 6.3 ± 0.065 |
4.036 ± 0.036 | 6.757 ± 0.056 |
1.556 ± 0.023 | 6.579 ± 0.046 |
6.793 ± 0.08 | 10.284 ± 0.075 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.776 ± 0.026 | 5.11 ± 0.068 |
3.841 ± 0.035 | 4.004 ± 0.038 |
4.917 ± 0.052 | 5.638 ± 0.063 |
5.527 ± 0.066 | 6.105 ± 0.052 |
1.051 ± 0.02 | 3.959 ± 0.05 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
