
Equine arteritis virus (strain Bucyrus) (EAV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Arnidovirineae; Arteriviridae; Equarterivirinae; Alphaarterivirus; Alphaarterivirus equid; Equine arteritis virus
Average proteome isoelectric point is 7.77
Get precalculated fractions of proteins
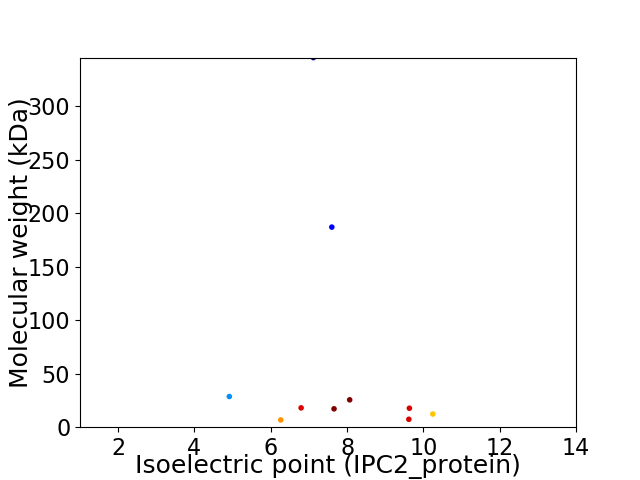
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
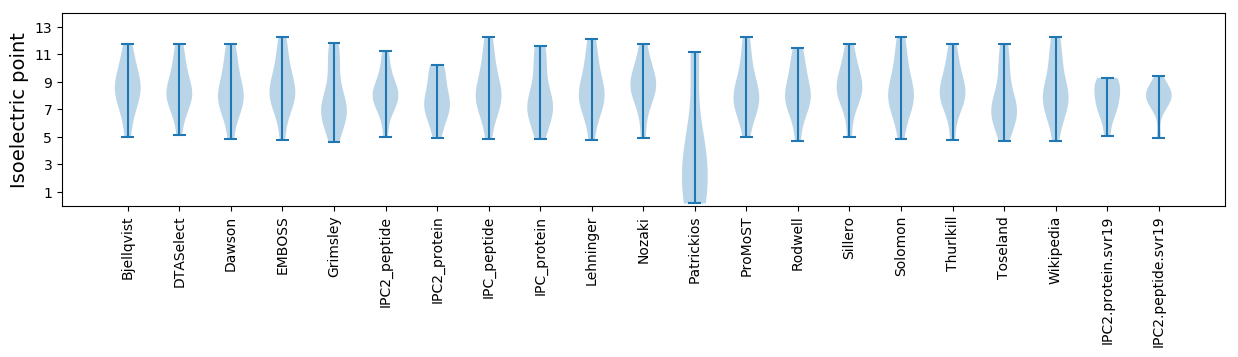
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q91DM1|E_EAVBU Envelope small membrane protein OS=Equine arteritis virus (strain Bucyrus) OX=299386 GN=GP2a PE=1 SV=1
MM1 pKa = 7.93LSMIVLLFLLWGAPSHH17 pKa = 6.92AYY19 pKa = 9.84FSYY22 pKa = 9.49YY23 pKa = 8.13TAQRR27 pKa = 11.84FTDD30 pKa = 4.11FTLCMLTDD38 pKa = 3.67RR39 pKa = 11.84GVIANLLRR47 pKa = 11.84YY48 pKa = 9.57DD49 pKa = 3.76EE50 pKa = 4.59HH51 pKa = 6.15TALYY55 pKa = 9.8NCSASKK61 pKa = 8.89TCWYY65 pKa = 8.61CTFLDD70 pKa = 3.81EE71 pKa = 5.32QIITFGTDD79 pKa = 2.76CDD81 pKa = 3.58DD82 pKa = 3.88TYY84 pKa = 11.63AVPVAEE90 pKa = 4.74VLEE93 pKa = 4.49QAHH96 pKa = 6.3GPYY99 pKa = 9.99SALFDD104 pKa = 4.38DD105 pKa = 5.07MPPFIYY111 pKa = 10.25YY112 pKa = 10.11GRR114 pKa = 11.84EE115 pKa = 3.52FGIVVLDD122 pKa = 3.16VFMFYY127 pKa = 10.43PVLVLFFLSVLPYY140 pKa = 9.36ATLILEE146 pKa = 4.33MCVSILFIIYY156 pKa = 9.84GIYY159 pKa = 10.04SGAYY163 pKa = 7.89LAMGIFAATLAIHH176 pKa = 6.54SIVVLRR182 pKa = 11.84QLLWLCLAWRR192 pKa = 11.84YY193 pKa = 10.1RR194 pKa = 11.84CTLHH198 pKa = 7.36ASFISAEE205 pKa = 4.12GKK207 pKa = 10.25VYY209 pKa = 10.23PVDD212 pKa = 4.17PGLPVAAVGNRR223 pKa = 11.84LLVPGRR229 pKa = 11.84PTIDD233 pKa = 3.26YY234 pKa = 10.46AVAYY238 pKa = 8.79GSKK241 pKa = 10.62VNLVRR246 pKa = 11.84LGAAEE251 pKa = 3.88VWEE254 pKa = 4.33PP255 pKa = 3.68
MM1 pKa = 7.93LSMIVLLFLLWGAPSHH17 pKa = 6.92AYY19 pKa = 9.84FSYY22 pKa = 9.49YY23 pKa = 8.13TAQRR27 pKa = 11.84FTDD30 pKa = 4.11FTLCMLTDD38 pKa = 3.67RR39 pKa = 11.84GVIANLLRR47 pKa = 11.84YY48 pKa = 9.57DD49 pKa = 3.76EE50 pKa = 4.59HH51 pKa = 6.15TALYY55 pKa = 9.8NCSASKK61 pKa = 8.89TCWYY65 pKa = 8.61CTFLDD70 pKa = 3.81EE71 pKa = 5.32QIITFGTDD79 pKa = 2.76CDD81 pKa = 3.58DD82 pKa = 3.88TYY84 pKa = 11.63AVPVAEE90 pKa = 4.74VLEE93 pKa = 4.49QAHH96 pKa = 6.3GPYY99 pKa = 9.99SALFDD104 pKa = 4.38DD105 pKa = 5.07MPPFIYY111 pKa = 10.25YY112 pKa = 10.11GRR114 pKa = 11.84EE115 pKa = 3.52FGIVVLDD122 pKa = 3.16VFMFYY127 pKa = 10.43PVLVLFFLSVLPYY140 pKa = 9.36ATLILEE146 pKa = 4.33MCVSILFIIYY156 pKa = 9.84GIYY159 pKa = 10.04SGAYY163 pKa = 7.89LAMGIFAATLAIHH176 pKa = 6.54SIVVLRR182 pKa = 11.84QLLWLCLAWRR192 pKa = 11.84YY193 pKa = 10.1RR194 pKa = 11.84CTLHH198 pKa = 7.36ASFISAEE205 pKa = 4.12GKK207 pKa = 10.25VYY209 pKa = 10.23PVDD212 pKa = 4.17PGLPVAAVGNRR223 pKa = 11.84LLVPGRR229 pKa = 11.84PTIDD233 pKa = 3.26YY234 pKa = 10.46AVAYY238 pKa = 8.79GSKK241 pKa = 10.62VNLVRR246 pKa = 11.84LGAAEE251 pKa = 3.88VWEE254 pKa = 4.33PP255 pKa = 3.68
Molecular weight: 28.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P19811-2|RPOA-2_EAVBU Isoform of P19811 Isoform Replicase polyprotein 1a of Replicase polyprotein 1ab OS=Equine arteritis virus (strain Bucyrus) OX=299386 GN=rep PE=1 SV=3
MM1 pKa = 7.22GLVWSLISNSIQTIIADD18 pKa = 3.94FAISVIDD25 pKa = 3.5AALFFLMLLALAVVTVFLFWLIVAIGRR52 pKa = 11.84SLVARR57 pKa = 11.84CSRR60 pKa = 11.84GARR63 pKa = 11.84YY64 pKa = 9.51RR65 pKa = 11.84PVV67 pKa = 2.74
MM1 pKa = 7.22GLVWSLISNSIQTIIADD18 pKa = 3.94FAISVIDD25 pKa = 3.5AALFFLMLLALAVVTVFLFWLIVAIGRR52 pKa = 11.84SLVARR57 pKa = 11.84CSRR60 pKa = 11.84GARR63 pKa = 11.84YY64 pKa = 9.51RR65 pKa = 11.84PVV67 pKa = 2.74
Molecular weight: 7.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6097 |
59 |
3175 |
609.7 |
66.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.578 ± 0.268 | 3.379 ± 0.306 |
4.658 ± 0.497 | 3.182 ± 0.533 |
4.478 ± 0.699 | 7.43 ± 0.724 |
2.263 ± 0.408 | 3.789 ± 0.781 |
3.477 ± 0.739 | 11.055 ± 1.01 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.919 ± 0.291 | 2.788 ± 0.185 |
5.872 ± 0.47 | 2.952 ± 0.245 |
5.363 ± 0.403 | 7.118 ± 0.484 |
6.282 ± 0.452 | 8.988 ± 0.465 |
1.673 ± 0.159 | 3.756 ± 0.543 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
