
Rhodoplanes sp. Z2-YC6860
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Hyphomicrobiaceae; Rhodoplanes; unclassified Rhodoplanes
Average proteome isoelectric point is 7.11
Get precalculated fractions of proteins
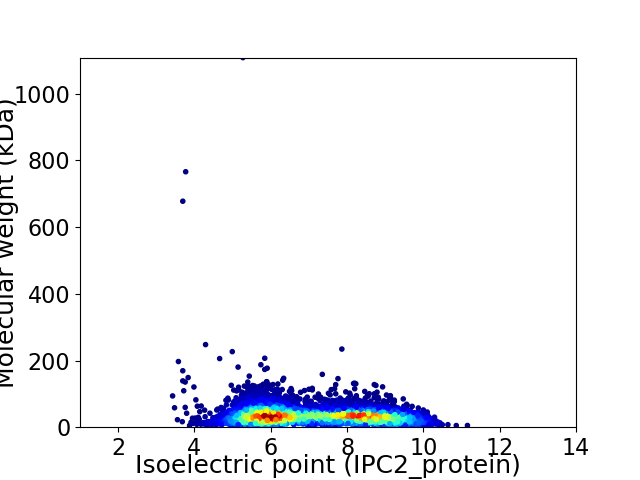
Virtual 2D-PAGE plot for 7007 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A127F2H4|A0A127F2H4_9RHIZ Transglutaminase family protein OS=Rhodoplanes sp. Z2-YC6860 OX=674703 GN=RHPLAN_53590 PE=4 SV=1
MM1 pKa = 7.41ATQSTGGGSTTSFGNAPQAVDD22 pKa = 3.69DD23 pKa = 4.73TFTSAQTGLTEE34 pKa = 4.92DD35 pKa = 3.84LLKK38 pKa = 10.84VVYY41 pKa = 10.19LDD43 pKa = 4.61VMANDD48 pKa = 4.2LGGNAKK54 pKa = 8.39TLFSIDD60 pKa = 3.0NGIYY64 pKa = 10.01TGTTVQSDD72 pKa = 4.59LLTQDD77 pKa = 3.25TARR80 pKa = 11.84AEE82 pKa = 4.14ATSADD87 pKa = 3.8TSLKK91 pKa = 9.86GAKK94 pKa = 8.94IWITSDD100 pKa = 3.07GKK102 pKa = 10.92VGYY105 pKa = 9.4DD106 pKa = 3.7ASTLSQSFKK115 pKa = 10.79DD116 pKa = 3.62QLSALHH122 pKa = 6.38AGEE125 pKa = 4.52YY126 pKa = 9.51LTDD129 pKa = 3.33TFTYY133 pKa = 10.22AIRR136 pKa = 11.84LGNGTLSWATATVQFAGANDD156 pKa = 3.84TATITASGNEE166 pKa = 4.08DD167 pKa = 3.49TTVVEE172 pKa = 5.15SGGTNNTTLGDD183 pKa = 3.9PSASGQLTVNDD194 pKa = 3.39VDD196 pKa = 3.69SGEE199 pKa = 4.21NHH201 pKa = 6.07FQTPSSLAGTYY212 pKa = 7.56GTYY215 pKa = 10.13TFNATSGVWGYY226 pKa = 7.48TLNQSLADD234 pKa = 4.22PLTAGQVVYY243 pKa = 10.97DD244 pKa = 3.61HH245 pKa = 7.16LIVKK249 pKa = 9.91SADD252 pKa = 3.01GTASHH257 pKa = 7.67DD258 pKa = 3.46IAVKK262 pKa = 9.05ITGSNDD268 pKa = 2.92SATITASSTEE278 pKa = 3.99DD279 pKa = 3.27TSVKK283 pKa = 10.1EE284 pKa = 4.06AGGVSNGAAGDD295 pKa = 3.99PSAHH299 pKa = 6.15GQLTVHH305 pKa = 6.84DD306 pKa = 4.15VDD308 pKa = 3.83SGEE311 pKa = 4.18NHH313 pKa = 6.06FQTPASLAGTYY324 pKa = 7.56GTYY327 pKa = 10.11TFNEE331 pKa = 4.36TSGAWGYY338 pKa = 7.48TLNQSLADD346 pKa = 4.02SLTDD350 pKa = 3.5GQVVHH355 pKa = 7.06DD356 pKa = 4.18HH357 pKa = 6.98LIVKK361 pKa = 10.02SADD364 pKa = 3.11NTTSYY369 pKa = 11.08DD370 pKa = 3.04IDD372 pKa = 3.74VTITGTNDD380 pKa = 2.91AATITASATEE390 pKa = 4.03DD391 pKa = 3.53TSVKK395 pKa = 10.09EE396 pKa = 4.06AGGVSNGTAGDD407 pKa = 4.0PSAHH411 pKa = 6.11GQLTVHH417 pKa = 6.84DD418 pKa = 4.15VDD420 pKa = 3.83SGEE423 pKa = 4.18NHH425 pKa = 6.06FQTPASLAGTYY436 pKa = 7.56GTYY439 pKa = 10.11TFNATTGAWSYY450 pKa = 9.67TLNQSIADD458 pKa = 3.9SLYY461 pKa = 10.5DD462 pKa = 3.41GQVAHH467 pKa = 7.2DD468 pKa = 3.93HH469 pKa = 6.53LIVRR473 pKa = 11.84SADD476 pKa = 3.06NTASYY481 pKa = 10.28DD482 pKa = 3.52INIDD486 pKa = 3.28ILGTNDD492 pKa = 3.37AAVLSSASVNLTEE505 pKa = 5.42GDD507 pKa = 3.5TAADD511 pKa = 3.28ISTSGTLTISDD522 pKa = 4.02VDD524 pKa = 3.93SDD526 pKa = 3.94PHH528 pKa = 7.41FVAQTDD534 pKa = 3.57TAGNYY539 pKa = 10.28GKK541 pKa = 10.63FSIDD545 pKa = 3.9ADD547 pKa = 4.3GAWSYY552 pKa = 11.37VADD555 pKa = 3.71SAHH558 pKa = 6.42NEE560 pKa = 3.9FAAGQHH566 pKa = 4.67YY567 pKa = 9.5TEE569 pKa = 4.84SFDD572 pKa = 4.0VVSADD577 pKa = 3.51GTHH580 pKa = 6.32TSVAIDD586 pKa = 3.13ILGTNDD592 pKa = 3.19AATVAFDD599 pKa = 3.73TVSFSDD605 pKa = 3.31TGSNDD610 pKa = 3.15GDD612 pKa = 3.64HH613 pKa = 6.58LTNNNAVTFSGSYY626 pKa = 10.42SDD628 pKa = 3.99VDD630 pKa = 3.64GTVTQIQVFNGSTSLGFATLATGTWTLNTALVDD663 pKa = 3.89GSYY666 pKa = 10.85NQLRR670 pKa = 11.84VLATDD675 pKa = 3.41NNRR678 pKa = 11.84ATTEE682 pKa = 4.07ATNPTQIIVDD692 pKa = 3.95TLNPTVSSVSINDD705 pKa = 3.47NQITDD710 pKa = 3.35GDD712 pKa = 3.96AAGIRR717 pKa = 11.84TATITFSEE725 pKa = 4.59AMDD728 pKa = 3.54QTSTPTISNSASSTLTSPTGGHH750 pKa = 5.61WVDD753 pKa = 3.3ATHH756 pKa = 6.64YY757 pKa = 9.99AINYY761 pKa = 5.83TAADD765 pKa = 3.51AGVTLNDD772 pKa = 2.73ITFNVSGAKK781 pKa = 10.09DD782 pKa = 3.3LAGNTQAAATNVSSGTSVDD801 pKa = 3.55TQNPTNTWSFSTSSKK816 pKa = 10.61AITVTASDD824 pKa = 3.34SSGIGSVTAVDD835 pKa = 3.72NTHH838 pKa = 7.22PGQGFAAAVNNGNGTWTIATSPSANINGDD867 pKa = 3.71SVTVTVTDD875 pKa = 3.71GAGNVTTSTHH885 pKa = 5.67NAPAGVAGSLINLGLTDD902 pKa = 4.53PSVDD906 pKa = 3.44HH907 pKa = 6.94IGAVSTVISGIPAGWILSEE926 pKa = 4.76GADD929 pKa = 3.26NGNGNWAVQTDD940 pKa = 3.75NLAALSIISSAAFTGALVLNVTEE963 pKa = 4.26TWTNADD969 pKa = 3.44GSNGTSIVSDD979 pKa = 3.56NVEE982 pKa = 4.03VFATGSPIFAWSGDD996 pKa = 3.5DD997 pKa = 3.93HH998 pKa = 6.67LTGSSGRR1005 pKa = 11.84DD1006 pKa = 2.94LFVFSQPIGADD1017 pKa = 3.34VIHH1020 pKa = 6.69NFDD1023 pKa = 3.87AAADD1027 pKa = 3.9QIDD1030 pKa = 4.64LIDD1033 pKa = 3.6YY1034 pKa = 9.59HH1035 pKa = 8.06SEE1037 pKa = 3.75SGATLTYY1044 pKa = 11.1ADD1046 pKa = 4.58LQSLLSQDD1054 pKa = 3.47GAGNAVISLGNGQSITLDD1072 pKa = 3.47GVHH1075 pKa = 6.83AADD1078 pKa = 3.78LSEE1081 pKa = 4.53ANFVFDD1087 pKa = 4.58VTPTLDD1093 pKa = 3.25NAGTMTIGDD1102 pKa = 4.06GAMLPLSGIINNTGTIEE1119 pKa = 4.2LDD1121 pKa = 3.47SAGSSTLLQLIQYY1134 pKa = 8.84GITLQGNGTVVLSDD1148 pKa = 3.19NDD1150 pKa = 3.79GNVISGSVQSVTLHH1164 pKa = 6.18NLDD1167 pKa = 3.5NTIEE1171 pKa = 4.31GAGQLGAGQLDD1182 pKa = 4.04LTNDD1186 pKa = 3.32GTIIANGSHH1195 pKa = 6.8ALVIDD1200 pKa = 3.64TGANVIEE1207 pKa = 4.13NAGMLEE1213 pKa = 3.96ATGTGGLVINSALEE1227 pKa = 3.82NDD1229 pKa = 4.12GLVWAHH1235 pKa = 6.77GGNITLSGAVTGSGSVQIDD1254 pKa = 3.36GNAKK1258 pKa = 9.3VSFGAVASVNTNIAADD1274 pKa = 3.71SASTVTVHH1282 pKa = 7.89DD1283 pKa = 4.05SFDD1286 pKa = 3.68FSGVISGFNGDD1297 pKa = 3.9DD1298 pKa = 3.82HH1299 pKa = 7.07MDD1301 pKa = 3.42LTDD1304 pKa = 3.3VMFSAGLTVNYY1315 pKa = 9.73AAAADD1320 pKa = 4.02GTGGVLQVSDD1330 pKa = 4.18GAHH1333 pKa = 5.67TANIALLGQYY1343 pKa = 9.23DD1344 pKa = 3.56ASGFRR1349 pKa = 11.84AGADD1353 pKa = 3.43SSSGTSITYY1362 pKa = 9.94DD1363 pKa = 3.21PHH1365 pKa = 5.61HH1366 pKa = 6.32TVV1368 pKa = 3.04
MM1 pKa = 7.41ATQSTGGGSTTSFGNAPQAVDD22 pKa = 3.69DD23 pKa = 4.73TFTSAQTGLTEE34 pKa = 4.92DD35 pKa = 3.84LLKK38 pKa = 10.84VVYY41 pKa = 10.19LDD43 pKa = 4.61VMANDD48 pKa = 4.2LGGNAKK54 pKa = 8.39TLFSIDD60 pKa = 3.0NGIYY64 pKa = 10.01TGTTVQSDD72 pKa = 4.59LLTQDD77 pKa = 3.25TARR80 pKa = 11.84AEE82 pKa = 4.14ATSADD87 pKa = 3.8TSLKK91 pKa = 9.86GAKK94 pKa = 8.94IWITSDD100 pKa = 3.07GKK102 pKa = 10.92VGYY105 pKa = 9.4DD106 pKa = 3.7ASTLSQSFKK115 pKa = 10.79DD116 pKa = 3.62QLSALHH122 pKa = 6.38AGEE125 pKa = 4.52YY126 pKa = 9.51LTDD129 pKa = 3.33TFTYY133 pKa = 10.22AIRR136 pKa = 11.84LGNGTLSWATATVQFAGANDD156 pKa = 3.84TATITASGNEE166 pKa = 4.08DD167 pKa = 3.49TTVVEE172 pKa = 5.15SGGTNNTTLGDD183 pKa = 3.9PSASGQLTVNDD194 pKa = 3.39VDD196 pKa = 3.69SGEE199 pKa = 4.21NHH201 pKa = 6.07FQTPSSLAGTYY212 pKa = 7.56GTYY215 pKa = 10.13TFNATSGVWGYY226 pKa = 7.48TLNQSLADD234 pKa = 4.22PLTAGQVVYY243 pKa = 10.97DD244 pKa = 3.61HH245 pKa = 7.16LIVKK249 pKa = 9.91SADD252 pKa = 3.01GTASHH257 pKa = 7.67DD258 pKa = 3.46IAVKK262 pKa = 9.05ITGSNDD268 pKa = 2.92SATITASSTEE278 pKa = 3.99DD279 pKa = 3.27TSVKK283 pKa = 10.1EE284 pKa = 4.06AGGVSNGAAGDD295 pKa = 3.99PSAHH299 pKa = 6.15GQLTVHH305 pKa = 6.84DD306 pKa = 4.15VDD308 pKa = 3.83SGEE311 pKa = 4.18NHH313 pKa = 6.06FQTPASLAGTYY324 pKa = 7.56GTYY327 pKa = 10.11TFNEE331 pKa = 4.36TSGAWGYY338 pKa = 7.48TLNQSLADD346 pKa = 4.02SLTDD350 pKa = 3.5GQVVHH355 pKa = 7.06DD356 pKa = 4.18HH357 pKa = 6.98LIVKK361 pKa = 10.02SADD364 pKa = 3.11NTTSYY369 pKa = 11.08DD370 pKa = 3.04IDD372 pKa = 3.74VTITGTNDD380 pKa = 2.91AATITASATEE390 pKa = 4.03DD391 pKa = 3.53TSVKK395 pKa = 10.09EE396 pKa = 4.06AGGVSNGTAGDD407 pKa = 4.0PSAHH411 pKa = 6.11GQLTVHH417 pKa = 6.84DD418 pKa = 4.15VDD420 pKa = 3.83SGEE423 pKa = 4.18NHH425 pKa = 6.06FQTPASLAGTYY436 pKa = 7.56GTYY439 pKa = 10.11TFNATTGAWSYY450 pKa = 9.67TLNQSIADD458 pKa = 3.9SLYY461 pKa = 10.5DD462 pKa = 3.41GQVAHH467 pKa = 7.2DD468 pKa = 3.93HH469 pKa = 6.53LIVRR473 pKa = 11.84SADD476 pKa = 3.06NTASYY481 pKa = 10.28DD482 pKa = 3.52INIDD486 pKa = 3.28ILGTNDD492 pKa = 3.37AAVLSSASVNLTEE505 pKa = 5.42GDD507 pKa = 3.5TAADD511 pKa = 3.28ISTSGTLTISDD522 pKa = 4.02VDD524 pKa = 3.93SDD526 pKa = 3.94PHH528 pKa = 7.41FVAQTDD534 pKa = 3.57TAGNYY539 pKa = 10.28GKK541 pKa = 10.63FSIDD545 pKa = 3.9ADD547 pKa = 4.3GAWSYY552 pKa = 11.37VADD555 pKa = 3.71SAHH558 pKa = 6.42NEE560 pKa = 3.9FAAGQHH566 pKa = 4.67YY567 pKa = 9.5TEE569 pKa = 4.84SFDD572 pKa = 4.0VVSADD577 pKa = 3.51GTHH580 pKa = 6.32TSVAIDD586 pKa = 3.13ILGTNDD592 pKa = 3.19AATVAFDD599 pKa = 3.73TVSFSDD605 pKa = 3.31TGSNDD610 pKa = 3.15GDD612 pKa = 3.64HH613 pKa = 6.58LTNNNAVTFSGSYY626 pKa = 10.42SDD628 pKa = 3.99VDD630 pKa = 3.64GTVTQIQVFNGSTSLGFATLATGTWTLNTALVDD663 pKa = 3.89GSYY666 pKa = 10.85NQLRR670 pKa = 11.84VLATDD675 pKa = 3.41NNRR678 pKa = 11.84ATTEE682 pKa = 4.07ATNPTQIIVDD692 pKa = 3.95TLNPTVSSVSINDD705 pKa = 3.47NQITDD710 pKa = 3.35GDD712 pKa = 3.96AAGIRR717 pKa = 11.84TATITFSEE725 pKa = 4.59AMDD728 pKa = 3.54QTSTPTISNSASSTLTSPTGGHH750 pKa = 5.61WVDD753 pKa = 3.3ATHH756 pKa = 6.64YY757 pKa = 9.99AINYY761 pKa = 5.83TAADD765 pKa = 3.51AGVTLNDD772 pKa = 2.73ITFNVSGAKK781 pKa = 10.09DD782 pKa = 3.3LAGNTQAAATNVSSGTSVDD801 pKa = 3.55TQNPTNTWSFSTSSKK816 pKa = 10.61AITVTASDD824 pKa = 3.34SSGIGSVTAVDD835 pKa = 3.72NTHH838 pKa = 7.22PGQGFAAAVNNGNGTWTIATSPSANINGDD867 pKa = 3.71SVTVTVTDD875 pKa = 3.71GAGNVTTSTHH885 pKa = 5.67NAPAGVAGSLINLGLTDD902 pKa = 4.53PSVDD906 pKa = 3.44HH907 pKa = 6.94IGAVSTVISGIPAGWILSEE926 pKa = 4.76GADD929 pKa = 3.26NGNGNWAVQTDD940 pKa = 3.75NLAALSIISSAAFTGALVLNVTEE963 pKa = 4.26TWTNADD969 pKa = 3.44GSNGTSIVSDD979 pKa = 3.56NVEE982 pKa = 4.03VFATGSPIFAWSGDD996 pKa = 3.5DD997 pKa = 3.93HH998 pKa = 6.67LTGSSGRR1005 pKa = 11.84DD1006 pKa = 2.94LFVFSQPIGADD1017 pKa = 3.34VIHH1020 pKa = 6.69NFDD1023 pKa = 3.87AAADD1027 pKa = 3.9QIDD1030 pKa = 4.64LIDD1033 pKa = 3.6YY1034 pKa = 9.59HH1035 pKa = 8.06SEE1037 pKa = 3.75SGATLTYY1044 pKa = 11.1ADD1046 pKa = 4.58LQSLLSQDD1054 pKa = 3.47GAGNAVISLGNGQSITLDD1072 pKa = 3.47GVHH1075 pKa = 6.83AADD1078 pKa = 3.78LSEE1081 pKa = 4.53ANFVFDD1087 pKa = 4.58VTPTLDD1093 pKa = 3.25NAGTMTIGDD1102 pKa = 4.06GAMLPLSGIINNTGTIEE1119 pKa = 4.2LDD1121 pKa = 3.47SAGSSTLLQLIQYY1134 pKa = 8.84GITLQGNGTVVLSDD1148 pKa = 3.19NDD1150 pKa = 3.79GNVISGSVQSVTLHH1164 pKa = 6.18NLDD1167 pKa = 3.5NTIEE1171 pKa = 4.31GAGQLGAGQLDD1182 pKa = 4.04LTNDD1186 pKa = 3.32GTIIANGSHH1195 pKa = 6.8ALVIDD1200 pKa = 3.64TGANVIEE1207 pKa = 4.13NAGMLEE1213 pKa = 3.96ATGTGGLVINSALEE1227 pKa = 3.82NDD1229 pKa = 4.12GLVWAHH1235 pKa = 6.77GGNITLSGAVTGSGSVQIDD1254 pKa = 3.36GNAKK1258 pKa = 9.3VSFGAVASVNTNIAADD1274 pKa = 3.71SASTVTVHH1282 pKa = 7.89DD1283 pKa = 4.05SFDD1286 pKa = 3.68FSGVISGFNGDD1297 pKa = 3.9DD1298 pKa = 3.82HH1299 pKa = 7.07MDD1301 pKa = 3.42LTDD1304 pKa = 3.3VMFSAGLTVNYY1315 pKa = 9.73AAAADD1320 pKa = 4.02GTGGVLQVSDD1330 pKa = 4.18GAHH1333 pKa = 5.67TANIALLGQYY1343 pKa = 9.23DD1344 pKa = 3.56ASGFRR1349 pKa = 11.84AGADD1353 pKa = 3.43SSSGTSITYY1362 pKa = 9.94DD1363 pKa = 3.21PHH1365 pKa = 5.61HH1366 pKa = 6.32TVV1368 pKa = 3.04
Molecular weight: 138.86 kDa
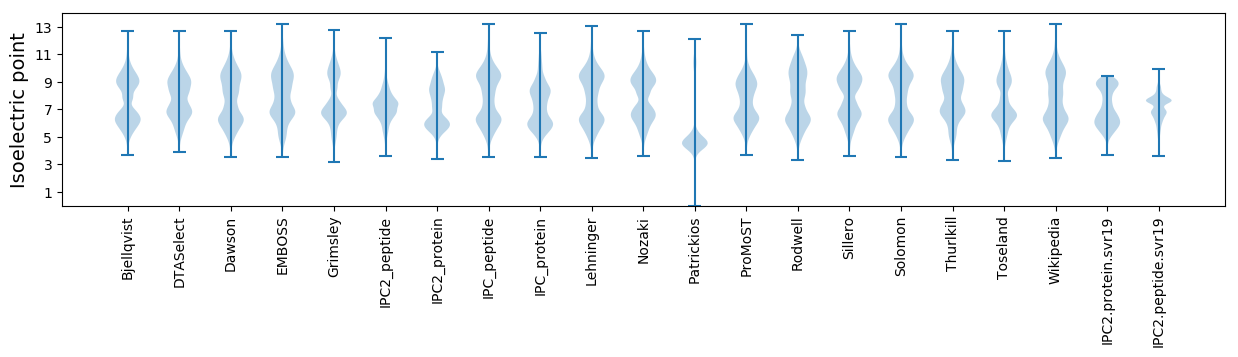
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A127F6W2|A0A127F6W2_9RHIZ Elongation factor P OS=Rhodoplanes sp. Z2-YC6860 OX=674703 GN=efp PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.65GFRR19 pKa = 11.84ARR21 pKa = 11.84LKK23 pKa = 7.72TTGGRR28 pKa = 11.84KK29 pKa = 8.99VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.65GFRR19 pKa = 11.84ARR21 pKa = 11.84LKK23 pKa = 7.72TTGGRR28 pKa = 11.84KK29 pKa = 8.99VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2322162 |
29 |
10376 |
331.4 |
35.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.2 ± 0.038 | 0.835 ± 0.009 |
5.432 ± 0.025 | 5.192 ± 0.031 |
3.764 ± 0.018 | 8.428 ± 0.032 |
2.043 ± 0.014 | 5.305 ± 0.02 |
4.035 ± 0.029 | 9.646 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.532 ± 0.016 | 2.86 ± 0.026 |
5.503 ± 0.023 | 3.24 ± 0.019 |
6.694 ± 0.039 | 5.504 ± 0.028 |
5.47 ± 0.048 | 7.65 ± 0.023 |
1.337 ± 0.011 | 2.33 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
