
Microviridae Fen7918_21
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.57
Get precalculated fractions of proteins
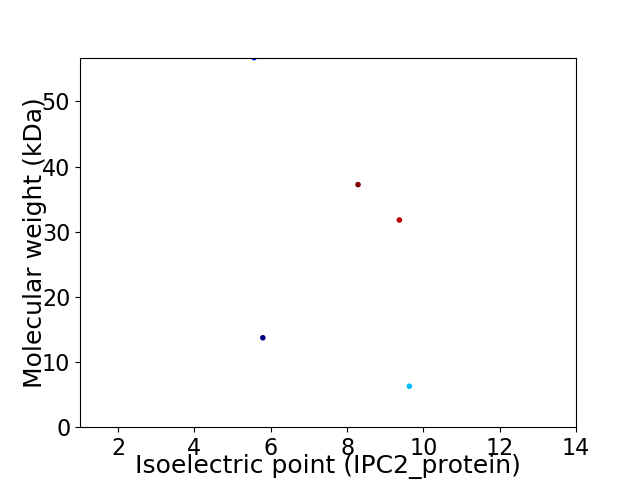
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
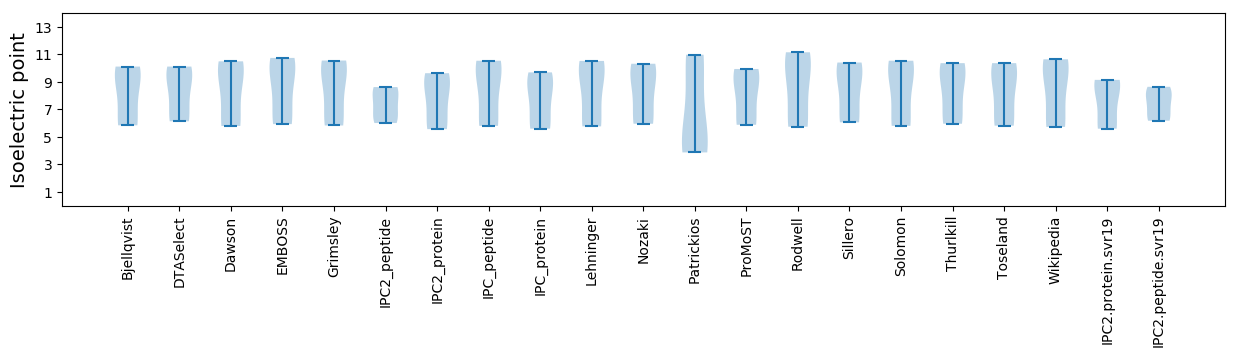
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G2UMI2|A0A0G2UMI2_9VIRU Uncharacterized protein OS=Microviridae Fen7918_21 OX=1655661 PE=4 SV=1
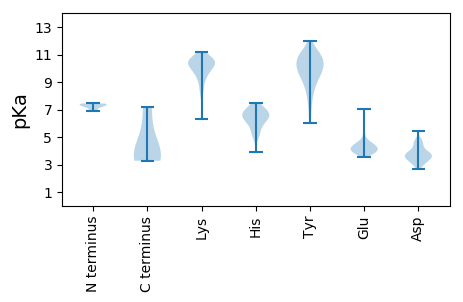
MM1 pKa = 7.36SRR3 pKa = 11.84NIFNSVKK10 pKa = 9.81MMRR13 pKa = 11.84PKK15 pKa = 10.84KK16 pKa = 10.22NVFDD20 pKa = 3.83LTHH23 pKa = 7.13DD24 pKa = 3.88VKK26 pKa = 10.37QTTRR30 pKa = 11.84FGEE33 pKa = 4.12LAPCLWLEE41 pKa = 4.46CVPGDD46 pKa = 3.66KK47 pKa = 10.61FKK49 pKa = 10.89IGAEE53 pKa = 4.0TLIRR57 pKa = 11.84FAPLISPVMHH67 pKa = 6.86RR68 pKa = 11.84MNASIHH74 pKa = 5.52YY75 pKa = 8.07FFVPYY80 pKa = 10.33RR81 pKa = 11.84ILWPNWEE88 pKa = 4.52DD89 pKa = 4.09YY90 pKa = 9.2ITNTSSVHH98 pKa = 5.35APPYY102 pKa = 10.1VDD104 pKa = 3.55YY105 pKa = 11.22SQIATSHH112 pKa = 5.0QRR114 pKa = 11.84LGDD117 pKa = 3.6FLGIPPQVQSGNGHH131 pKa = 6.47PKK133 pKa = 9.68VSAFPFAAYY142 pKa = 10.26LKK144 pKa = 10.3IYY146 pKa = 10.07DD147 pKa = 4.76DD148 pKa = 4.24YY149 pKa = 12.01YY150 pKa = 11.32RR151 pKa = 11.84DD152 pKa = 3.5QNLIQPAWSPLTDD165 pKa = 3.65GLQVANLVDD174 pKa = 4.61LVTMRR179 pKa = 11.84NRR181 pKa = 11.84AWEE184 pKa = 3.98HH185 pKa = 6.98DD186 pKa = 4.46YY187 pKa = 9.61FTASLPWAQKK197 pKa = 9.17GAQVEE202 pKa = 4.42IPLGDD207 pKa = 3.24ITLGQPAGGPNQFLRR222 pKa = 11.84RR223 pKa = 11.84GIDD226 pKa = 3.23GSTEE230 pKa = 3.85GNIAALGTDD239 pKa = 3.45TPGFLVDD246 pKa = 4.23AAGQVPVNIDD256 pKa = 3.49PNGSLINEE264 pKa = 3.97ATTINDD270 pKa = 2.96LRR272 pKa = 11.84RR273 pKa = 11.84AFRR276 pKa = 11.84LQEE279 pKa = 3.89WLEE282 pKa = 3.97KK283 pKa = 10.14NARR286 pKa = 11.84AGTRR290 pKa = 11.84YY291 pKa = 9.67IEE293 pKa = 4.73NILAHH298 pKa = 6.4FGVNSSDD305 pKa = 3.63KK306 pKa = 10.04RR307 pKa = 11.84LQRR310 pKa = 11.84PEE312 pKa = 4.34YY313 pKa = 9.97ICGLKK318 pKa = 9.33TPVVISEE325 pKa = 4.11VLNTTGAPDD334 pKa = 3.88GLAQGNMAGHH344 pKa = 7.44GIAAHH349 pKa = 5.14SAKK352 pKa = 10.47DD353 pKa = 3.51GSYY356 pKa = 9.86SCEE359 pKa = 3.7EE360 pKa = 4.31HH361 pKa = 6.53GLIMGVMSIMPNTAYY376 pKa = 10.25QQGIHH381 pKa = 6.38KK382 pKa = 9.67SWSKK386 pKa = 10.76FDD388 pKa = 3.4QLDD391 pKa = 4.0YY392 pKa = 10.84FWPTFAHH399 pKa = 6.84LGEE402 pKa = 4.2QEE404 pKa = 4.19VLNQEE409 pKa = 4.6VYY411 pKa = 10.62AQSSDD416 pKa = 3.26PGGTFGYY423 pKa = 9.46VPRR426 pKa = 11.84YY427 pKa = 9.67SEE429 pKa = 4.3YY430 pKa = 10.57KK431 pKa = 10.27FMPNRR436 pKa = 11.84VATSFRR442 pKa = 11.84DD443 pKa = 3.8TLAFWHH449 pKa = 6.52MGRR452 pKa = 11.84IFATEE457 pKa = 3.55PALNQDD463 pKa = 3.82FVEE466 pKa = 4.57CTTDD470 pKa = 3.0NRR472 pKa = 11.84IFAVTDD478 pKa = 3.73PDD480 pKa = 3.55VDD482 pKa = 4.7NIYY485 pKa = 10.94CDD487 pKa = 2.99ILNRR491 pKa = 11.84ITAIRR496 pKa = 11.84PMPKK500 pKa = 9.88YY501 pKa = 7.48GTPMII506 pKa = 4.76
MM1 pKa = 7.36SRR3 pKa = 11.84NIFNSVKK10 pKa = 9.81MMRR13 pKa = 11.84PKK15 pKa = 10.84KK16 pKa = 10.22NVFDD20 pKa = 3.83LTHH23 pKa = 7.13DD24 pKa = 3.88VKK26 pKa = 10.37QTTRR30 pKa = 11.84FGEE33 pKa = 4.12LAPCLWLEE41 pKa = 4.46CVPGDD46 pKa = 3.66KK47 pKa = 10.61FKK49 pKa = 10.89IGAEE53 pKa = 4.0TLIRR57 pKa = 11.84FAPLISPVMHH67 pKa = 6.86RR68 pKa = 11.84MNASIHH74 pKa = 5.52YY75 pKa = 8.07FFVPYY80 pKa = 10.33RR81 pKa = 11.84ILWPNWEE88 pKa = 4.52DD89 pKa = 4.09YY90 pKa = 9.2ITNTSSVHH98 pKa = 5.35APPYY102 pKa = 10.1VDD104 pKa = 3.55YY105 pKa = 11.22SQIATSHH112 pKa = 5.0QRR114 pKa = 11.84LGDD117 pKa = 3.6FLGIPPQVQSGNGHH131 pKa = 6.47PKK133 pKa = 9.68VSAFPFAAYY142 pKa = 10.26LKK144 pKa = 10.3IYY146 pKa = 10.07DD147 pKa = 4.76DD148 pKa = 4.24YY149 pKa = 12.01YY150 pKa = 11.32RR151 pKa = 11.84DD152 pKa = 3.5QNLIQPAWSPLTDD165 pKa = 3.65GLQVANLVDD174 pKa = 4.61LVTMRR179 pKa = 11.84NRR181 pKa = 11.84AWEE184 pKa = 3.98HH185 pKa = 6.98DD186 pKa = 4.46YY187 pKa = 9.61FTASLPWAQKK197 pKa = 9.17GAQVEE202 pKa = 4.42IPLGDD207 pKa = 3.24ITLGQPAGGPNQFLRR222 pKa = 11.84RR223 pKa = 11.84GIDD226 pKa = 3.23GSTEE230 pKa = 3.85GNIAALGTDD239 pKa = 3.45TPGFLVDD246 pKa = 4.23AAGQVPVNIDD256 pKa = 3.49PNGSLINEE264 pKa = 3.97ATTINDD270 pKa = 2.96LRR272 pKa = 11.84RR273 pKa = 11.84AFRR276 pKa = 11.84LQEE279 pKa = 3.89WLEE282 pKa = 3.97KK283 pKa = 10.14NARR286 pKa = 11.84AGTRR290 pKa = 11.84YY291 pKa = 9.67IEE293 pKa = 4.73NILAHH298 pKa = 6.4FGVNSSDD305 pKa = 3.63KK306 pKa = 10.04RR307 pKa = 11.84LQRR310 pKa = 11.84PEE312 pKa = 4.34YY313 pKa = 9.97ICGLKK318 pKa = 9.33TPVVISEE325 pKa = 4.11VLNTTGAPDD334 pKa = 3.88GLAQGNMAGHH344 pKa = 7.44GIAAHH349 pKa = 5.14SAKK352 pKa = 10.47DD353 pKa = 3.51GSYY356 pKa = 9.86SCEE359 pKa = 3.7EE360 pKa = 4.31HH361 pKa = 6.53GLIMGVMSIMPNTAYY376 pKa = 10.25QQGIHH381 pKa = 6.38KK382 pKa = 9.67SWSKK386 pKa = 10.76FDD388 pKa = 3.4QLDD391 pKa = 4.0YY392 pKa = 10.84FWPTFAHH399 pKa = 6.84LGEE402 pKa = 4.2QEE404 pKa = 4.19VLNQEE409 pKa = 4.6VYY411 pKa = 10.62AQSSDD416 pKa = 3.26PGGTFGYY423 pKa = 9.46VPRR426 pKa = 11.84YY427 pKa = 9.67SEE429 pKa = 4.3YY430 pKa = 10.57KK431 pKa = 10.27FMPNRR436 pKa = 11.84VATSFRR442 pKa = 11.84DD443 pKa = 3.8TLAFWHH449 pKa = 6.52MGRR452 pKa = 11.84IFATEE457 pKa = 3.55PALNQDD463 pKa = 3.82FVEE466 pKa = 4.57CTTDD470 pKa = 3.0NRR472 pKa = 11.84IFAVTDD478 pKa = 3.73PDD480 pKa = 3.55VDD482 pKa = 4.7NIYY485 pKa = 10.94CDD487 pKa = 2.99ILNRR491 pKa = 11.84ITAIRR496 pKa = 11.84PMPKK500 pKa = 9.88YY501 pKa = 7.48GTPMII506 pKa = 4.76
Molecular weight: 56.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G2UMI2|A0A0G2UMI2_9VIRU Uncharacterized protein OS=Microviridae Fen7918_21 OX=1655661 PE=4 SV=1
MM1 pKa = 6.9TQINTYY7 pKa = 9.47AQNATIWDD15 pKa = 4.26LVAILASDD23 pKa = 4.89LLTIAIIYY31 pKa = 7.6FIKK34 pKa = 10.14KK35 pKa = 8.68HH36 pKa = 5.94KK37 pKa = 10.41KK38 pKa = 9.55HH39 pKa = 6.04GLQKK43 pKa = 9.64NTWQAFLRR51 pKa = 11.84SRR53 pKa = 11.84PP54 pKa = 3.57
MM1 pKa = 6.9TQINTYY7 pKa = 9.47AQNATIWDD15 pKa = 4.26LVAILASDD23 pKa = 4.89LLTIAIIYY31 pKa = 7.6FIKK34 pKa = 10.14KK35 pKa = 8.68HH36 pKa = 5.94KK37 pKa = 10.41KK38 pKa = 9.55HH39 pKa = 6.04GLQKK43 pKa = 9.64NTWQAFLRR51 pKa = 11.84SRR53 pKa = 11.84PP54 pKa = 3.57
Molecular weight: 6.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1293 |
54 |
506 |
258.6 |
29.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.425 ± 0.961 | 0.928 ± 0.349 |
5.182 ± 0.612 | 4.099 ± 0.568 |
3.635 ± 0.644 | 6.497 ± 0.792 |
2.552 ± 0.721 | 6.11 ± 1.091 |
6.032 ± 1.605 | 8.507 ± 0.585 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.475 ± 0.279 | 6.961 ± 0.962 |
5.182 ± 0.892 | 6.342 ± 1.549 |
4.95 ± 0.704 | 6.11 ± 0.46 |
7.115 ± 0.515 | 4.408 ± 0.831 |
1.779 ± 0.211 | 3.712 ± 0.456 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
