
Breda virus 1 (BRV-1)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Tornidovirineae; Tobaniviridae; Torovirinae; Torovirus; Renitovirus; Bovine torovirus; Breda virus
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
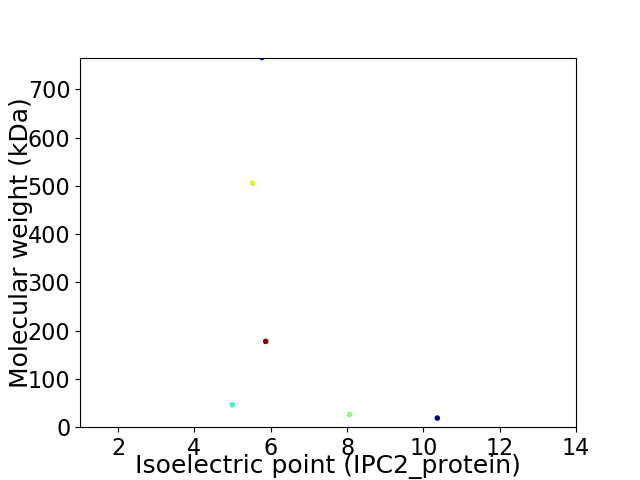
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
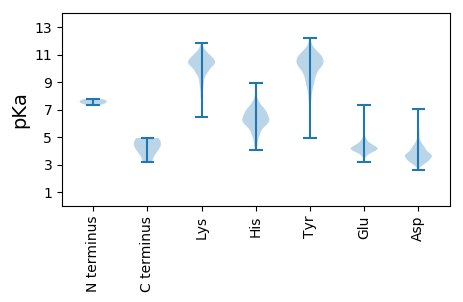
>sp|P0C6F4|R1A_BRV1 Replicase polyprotein 1a OS=Breda virus 1 OX=360393 GN=1a PE=3 SV=1
MM1 pKa = 7.64LSLILFFPSFAFAATPVTPYY21 pKa = 10.05YY22 pKa = 10.85GPGHH26 pKa = 6.45ITFDD30 pKa = 2.96WCGFGDD36 pKa = 4.6SRR38 pKa = 11.84SDD40 pKa = 3.4CTNPQSPMSLDD51 pKa = 3.52IPQQLCPKK59 pKa = 9.3FSSKK63 pKa = 10.56SSSSMFLSLHH73 pKa = 5.92WNNHH77 pKa = 4.49SSFVSYY83 pKa = 10.79DD84 pKa = 3.41YY85 pKa = 10.99FNCGVEE91 pKa = 4.1KK92 pKa = 10.87VFYY95 pKa = 10.1EE96 pKa = 4.4GVNFSPRR103 pKa = 11.84KK104 pKa = 9.63QYY106 pKa = 10.99SCWDD110 pKa = 3.52EE111 pKa = 5.57GVDD114 pKa = 3.97GWIEE118 pKa = 4.07LKK120 pKa = 10.03TRR122 pKa = 11.84FYY124 pKa = 10.93TKK126 pKa = 9.92LYY128 pKa = 10.95QMATTSRR135 pKa = 11.84CIKK138 pKa = 10.23LIQLQAPSSLPTLQAGVCRR157 pKa = 11.84TNKK160 pKa = 9.59QLPDD164 pKa = 3.62NPRR167 pKa = 11.84LALLSDD173 pKa = 4.13TVPTSVQFVLPGSSGTTICTKK194 pKa = 10.43HH195 pKa = 6.21LVPFCYY201 pKa = 10.16LNHH204 pKa = 6.67GCFTTGGSCLPFGVSYY220 pKa = 11.25VSDD223 pKa = 3.46SFYY226 pKa = 11.05YY227 pKa = 10.33GYY229 pKa = 11.38YY230 pKa = 10.49DD231 pKa = 3.59ATPQIGSTEE240 pKa = 3.98SHH242 pKa = 7.45DD243 pKa = 4.07YY244 pKa = 10.11VCDD247 pKa = 3.7YY248 pKa = 11.57LFMEE252 pKa = 5.47PGTYY256 pKa = 9.82NASTVGKK263 pKa = 9.6FLVYY267 pKa = 8.18PTKK270 pKa = 10.33SYY272 pKa = 11.54CMDD275 pKa = 3.38TMNITVPVQAVQSIWSEE292 pKa = 4.19QYY294 pKa = 11.3ASDD297 pKa = 4.15DD298 pKa = 5.34AIGQACKK305 pKa = 10.37APYY308 pKa = 10.08CIFYY312 pKa = 11.06NKK314 pKa = 6.93TTPYY318 pKa = 9.94TVTNGSDD325 pKa = 3.28ANHH328 pKa = 7.09GDD330 pKa = 3.56DD331 pKa = 4.3EE332 pKa = 4.86VRR334 pKa = 11.84MMMQGLLRR342 pKa = 11.84NSSCISPQGSTPLALYY358 pKa = 7.87STEE361 pKa = 4.51MIYY364 pKa = 10.68EE365 pKa = 4.21PNYY368 pKa = 9.67GSCPQFYY375 pKa = 10.87KK376 pKa = 11.08LFDD379 pKa = 3.36TSGNEE384 pKa = 4.06NIDD387 pKa = 4.31VISSSYY393 pKa = 10.38FVATWVLLVVVVILIFVIISFFCC416 pKa = 4.13
MM1 pKa = 7.64LSLILFFPSFAFAATPVTPYY21 pKa = 10.05YY22 pKa = 10.85GPGHH26 pKa = 6.45ITFDD30 pKa = 2.96WCGFGDD36 pKa = 4.6SRR38 pKa = 11.84SDD40 pKa = 3.4CTNPQSPMSLDD51 pKa = 3.52IPQQLCPKK59 pKa = 9.3FSSKK63 pKa = 10.56SSSSMFLSLHH73 pKa = 5.92WNNHH77 pKa = 4.49SSFVSYY83 pKa = 10.79DD84 pKa = 3.41YY85 pKa = 10.99FNCGVEE91 pKa = 4.1KK92 pKa = 10.87VFYY95 pKa = 10.1EE96 pKa = 4.4GVNFSPRR103 pKa = 11.84KK104 pKa = 9.63QYY106 pKa = 10.99SCWDD110 pKa = 3.52EE111 pKa = 5.57GVDD114 pKa = 3.97GWIEE118 pKa = 4.07LKK120 pKa = 10.03TRR122 pKa = 11.84FYY124 pKa = 10.93TKK126 pKa = 9.92LYY128 pKa = 10.95QMATTSRR135 pKa = 11.84CIKK138 pKa = 10.23LIQLQAPSSLPTLQAGVCRR157 pKa = 11.84TNKK160 pKa = 9.59QLPDD164 pKa = 3.62NPRR167 pKa = 11.84LALLSDD173 pKa = 4.13TVPTSVQFVLPGSSGTTICTKK194 pKa = 10.43HH195 pKa = 6.21LVPFCYY201 pKa = 10.16LNHH204 pKa = 6.67GCFTTGGSCLPFGVSYY220 pKa = 11.25VSDD223 pKa = 3.46SFYY226 pKa = 11.05YY227 pKa = 10.33GYY229 pKa = 11.38YY230 pKa = 10.49DD231 pKa = 3.59ATPQIGSTEE240 pKa = 3.98SHH242 pKa = 7.45DD243 pKa = 4.07YY244 pKa = 10.11VCDD247 pKa = 3.7YY248 pKa = 11.57LFMEE252 pKa = 5.47PGTYY256 pKa = 9.82NASTVGKK263 pKa = 9.6FLVYY267 pKa = 8.18PTKK270 pKa = 10.33SYY272 pKa = 11.54CMDD275 pKa = 3.38TMNITVPVQAVQSIWSEE292 pKa = 4.19QYY294 pKa = 11.3ASDD297 pKa = 4.15DD298 pKa = 5.34AIGQACKK305 pKa = 10.37APYY308 pKa = 10.08CIFYY312 pKa = 11.06NKK314 pKa = 6.93TTPYY318 pKa = 9.94TVTNGSDD325 pKa = 3.28ANHH328 pKa = 7.09GDD330 pKa = 3.56DD331 pKa = 4.3EE332 pKa = 4.86VRR334 pKa = 11.84MMMQGLLRR342 pKa = 11.84NSSCISPQGSTPLALYY358 pKa = 7.87STEE361 pKa = 4.51MIYY364 pKa = 10.68EE365 pKa = 4.21PNYY368 pKa = 9.67GSCPQFYY375 pKa = 10.87KK376 pKa = 11.08LFDD379 pKa = 3.36TSGNEE384 pKa = 4.06NIDD387 pKa = 4.31VISSSYY393 pKa = 10.38FVATWVLLVVVVILIFVIISFFCC416 pKa = 4.13
Molecular weight: 46.44 kDa
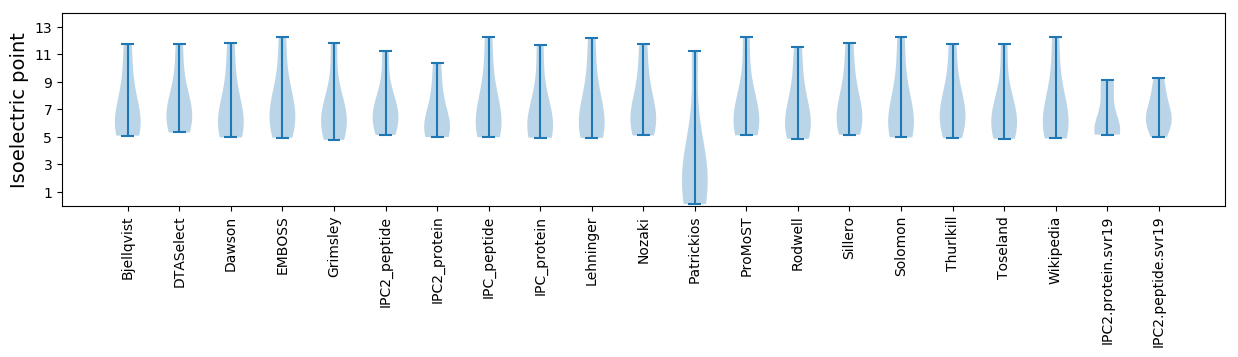
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P0C0V9|HEMA_BRV1 Hemagglutinin-esterase OS=Breda virus 1 OX=360393 GN=HE PE=1 SV=1
MM1 pKa = 7.76NSMLNPNAVPCQPSPQVVAIPMQYY25 pKa = 10.46PSGFSPGFRR34 pKa = 11.84RR35 pKa = 11.84QRR37 pKa = 11.84NPGFRR42 pKa = 11.84PMFNRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84NNNGNQNRR57 pKa = 11.84GRR59 pKa = 11.84QNRR62 pKa = 11.84QRR64 pKa = 11.84VQNNNRR70 pKa = 11.84GNIRR74 pKa = 11.84NRR76 pKa = 11.84QNNGQRR82 pKa = 11.84GNRR85 pKa = 11.84RR86 pKa = 11.84QYY88 pKa = 10.08NQPSPNVPFEE98 pKa = 4.02QQLLMMANEE107 pKa = 3.96TAYY110 pKa = 10.53AATYY114 pKa = 8.79PPEE117 pKa = 4.19MQNVAPTKK125 pKa = 9.86LVKK128 pKa = 9.38IAKK131 pKa = 8.54RR132 pKa = 11.84AAMQIVSGHH141 pKa = 5.28ATVEE145 pKa = 4.05ISNGTEE151 pKa = 3.76DD152 pKa = 3.39SNKK155 pKa = 9.84RR156 pKa = 11.84VATFTIKK163 pKa = 10.74VVMNN167 pKa = 4.55
MM1 pKa = 7.76NSMLNPNAVPCQPSPQVVAIPMQYY25 pKa = 10.46PSGFSPGFRR34 pKa = 11.84RR35 pKa = 11.84QRR37 pKa = 11.84NPGFRR42 pKa = 11.84PMFNRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84NNNGNQNRR57 pKa = 11.84GRR59 pKa = 11.84QNRR62 pKa = 11.84QRR64 pKa = 11.84VQNNNRR70 pKa = 11.84GNIRR74 pKa = 11.84NRR76 pKa = 11.84QNNGQRR82 pKa = 11.84GNRR85 pKa = 11.84RR86 pKa = 11.84QYY88 pKa = 10.08NQPSPNVPFEE98 pKa = 4.02QQLLMMANEE107 pKa = 3.96TAYY110 pKa = 10.53AATYY114 pKa = 8.79PPEE117 pKa = 4.19MQNVAPTKK125 pKa = 9.86LVKK128 pKa = 9.38IAKK131 pKa = 8.54RR132 pKa = 11.84AAMQIVSGHH141 pKa = 5.28ATVEE145 pKa = 4.05ISNGTEE151 pKa = 3.76DD152 pKa = 3.39SNKK155 pKa = 9.84RR156 pKa = 11.84VATFTIKK163 pKa = 10.74VVMNN167 pKa = 4.55
Molecular weight: 18.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13577 |
167 |
6733 |
2262.8 |
256.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.81 ± 0.617 | 3.256 ± 0.239 |
5.266 ± 0.68 | 4.309 ± 0.459 |
6.194 ± 0.329 | 5.075 ± 0.133 |
2.21 ± 0.233 | 4.456 ± 0.463 |
4.92 ± 0.592 | 9.951 ± 0.491 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.114 ± 0.203 | 4.382 ± 0.446 |
4.935 ± 0.339 | 4.611 ± 0.321 |
3.609 ± 0.266 | 7.785 ± 0.658 |
5.738 ± 0.603 | 9.789 ± 0.806 |
1.451 ± 0.171 | 5.112 ± 0.341 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
