
Microbacterium sediminis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
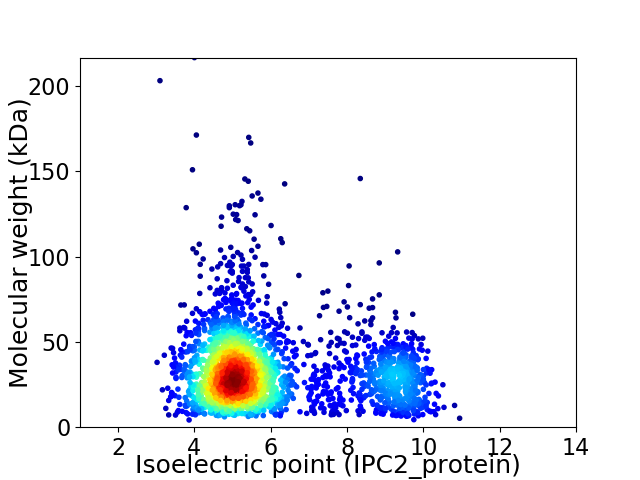
Virtual 2D-PAGE plot for 2529 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B9NB39|A0A1B9NB39_9MICO Histidinol-phosphate aminotransferase OS=Microbacterium sediminis OX=904291 GN=hisC PE=3 SV=1
MM1 pKa = 7.18NVRR4 pKa = 11.84RR5 pKa = 11.84SWLAAAAVTLSAGLALTGCAGAGNTGNTNGGSGDD39 pKa = 3.78EE40 pKa = 4.09NTIVWWHH47 pKa = 5.99NSNTGEE53 pKa = 3.97GKK55 pKa = 10.27EE56 pKa = 4.34YY57 pKa = 10.21YY58 pKa = 10.19DD59 pKa = 3.62QVAADD64 pKa = 4.67FEE66 pKa = 4.52AAHH69 pKa = 6.54EE70 pKa = 4.41GVTVQVEE77 pKa = 4.35AMQHH81 pKa = 5.21EE82 pKa = 4.71DD83 pKa = 3.72MLTKK87 pKa = 10.54LQAAFQGGDD96 pKa = 3.0AAQIPDD102 pKa = 3.54VYY104 pKa = 9.67MSRR107 pKa = 11.84GGGEE111 pKa = 4.03LKK113 pKa = 10.68SEE115 pKa = 4.3VEE117 pKa = 4.15GGVVRR122 pKa = 11.84DD123 pKa = 3.94LTEE126 pKa = 4.37DD127 pKa = 3.45AADD130 pKa = 4.1TISTIEE136 pKa = 4.38SFTEE140 pKa = 4.12QYY142 pKa = 10.34TVDD145 pKa = 3.8DD146 pKa = 4.01KK147 pKa = 11.97VYY149 pKa = 10.42ALPYY153 pKa = 10.03SIGIVGFWYY162 pKa = 10.75NKK164 pKa = 10.39DD165 pKa = 3.07LFAAAGIDD173 pKa = 3.51EE174 pKa = 4.59VSPNPTWEE182 pKa = 4.02EE183 pKa = 3.5FTGYY187 pKa = 10.11VDD189 pKa = 3.45KK190 pKa = 11.47LKK192 pKa = 10.91AAGTAPIAVGAGDD205 pKa = 3.89KK206 pKa = 10.12WPAAHH211 pKa = 5.66YY212 pKa = 8.9WYY214 pKa = 10.53YY215 pKa = 11.12NVVRR219 pKa = 11.84EE220 pKa = 4.14CEE222 pKa = 4.05YY223 pKa = 11.38DD224 pKa = 3.7VVEE227 pKa = 4.32AAIEE231 pKa = 4.22SGDD234 pKa = 3.73YY235 pKa = 10.77SDD237 pKa = 5.32EE238 pKa = 4.45CFLTAGEE245 pKa = 4.37HH246 pKa = 6.37LEE248 pKa = 4.09EE249 pKa = 4.31TLALEE254 pKa = 4.27PFNAGFLSTPAQSGPTSASGLLATGKK280 pKa = 10.61VGMEE284 pKa = 4.18LAGHH288 pKa = 6.36WEE290 pKa = 3.98PGVAGGLTEE299 pKa = 4.3NGEE302 pKa = 4.4VPPFLGWFAYY312 pKa = 7.59PTFDD316 pKa = 3.93GQGGAPDD323 pKa = 3.93DD324 pKa = 3.85QMGGGDD330 pKa = 3.61AWSVSTGAPDD340 pKa = 3.15IAVEE344 pKa = 4.02FAEE347 pKa = 4.37YY348 pKa = 10.69LLSDD352 pKa = 3.99EE353 pKa = 4.41VQIGFAEE360 pKa = 4.59LDD362 pKa = 3.24MGLPTNPAATDD373 pKa = 3.44SLANEE378 pKa = 4.35TLAQLIPVRR387 pKa = 11.84DD388 pKa = 4.04GGGKK392 pKa = 6.82TQLYY396 pKa = 10.19LDD398 pKa = 3.84TRR400 pKa = 11.84LGQSVGNALNDD411 pKa = 4.58AIALMFAGQAGPEE424 pKa = 4.48DD425 pKa = 3.47IVEE428 pKa = 4.34ALEE431 pKa = 4.24SASAMGG437 pKa = 3.92
MM1 pKa = 7.18NVRR4 pKa = 11.84RR5 pKa = 11.84SWLAAAAVTLSAGLALTGCAGAGNTGNTNGGSGDD39 pKa = 3.78EE40 pKa = 4.09NTIVWWHH47 pKa = 5.99NSNTGEE53 pKa = 3.97GKK55 pKa = 10.27EE56 pKa = 4.34YY57 pKa = 10.21YY58 pKa = 10.19DD59 pKa = 3.62QVAADD64 pKa = 4.67FEE66 pKa = 4.52AAHH69 pKa = 6.54EE70 pKa = 4.41GVTVQVEE77 pKa = 4.35AMQHH81 pKa = 5.21EE82 pKa = 4.71DD83 pKa = 3.72MLTKK87 pKa = 10.54LQAAFQGGDD96 pKa = 3.0AAQIPDD102 pKa = 3.54VYY104 pKa = 9.67MSRR107 pKa = 11.84GGGEE111 pKa = 4.03LKK113 pKa = 10.68SEE115 pKa = 4.3VEE117 pKa = 4.15GGVVRR122 pKa = 11.84DD123 pKa = 3.94LTEE126 pKa = 4.37DD127 pKa = 3.45AADD130 pKa = 4.1TISTIEE136 pKa = 4.38SFTEE140 pKa = 4.12QYY142 pKa = 10.34TVDD145 pKa = 3.8DD146 pKa = 4.01KK147 pKa = 11.97VYY149 pKa = 10.42ALPYY153 pKa = 10.03SIGIVGFWYY162 pKa = 10.75NKK164 pKa = 10.39DD165 pKa = 3.07LFAAAGIDD173 pKa = 3.51EE174 pKa = 4.59VSPNPTWEE182 pKa = 4.02EE183 pKa = 3.5FTGYY187 pKa = 10.11VDD189 pKa = 3.45KK190 pKa = 11.47LKK192 pKa = 10.91AAGTAPIAVGAGDD205 pKa = 3.89KK206 pKa = 10.12WPAAHH211 pKa = 5.66YY212 pKa = 8.9WYY214 pKa = 10.53YY215 pKa = 11.12NVVRR219 pKa = 11.84EE220 pKa = 4.14CEE222 pKa = 4.05YY223 pKa = 11.38DD224 pKa = 3.7VVEE227 pKa = 4.32AAIEE231 pKa = 4.22SGDD234 pKa = 3.73YY235 pKa = 10.77SDD237 pKa = 5.32EE238 pKa = 4.45CFLTAGEE245 pKa = 4.37HH246 pKa = 6.37LEE248 pKa = 4.09EE249 pKa = 4.31TLALEE254 pKa = 4.27PFNAGFLSTPAQSGPTSASGLLATGKK280 pKa = 10.61VGMEE284 pKa = 4.18LAGHH288 pKa = 6.36WEE290 pKa = 3.98PGVAGGLTEE299 pKa = 4.3NGEE302 pKa = 4.4VPPFLGWFAYY312 pKa = 7.59PTFDD316 pKa = 3.93GQGGAPDD323 pKa = 3.93DD324 pKa = 3.85QMGGGDD330 pKa = 3.61AWSVSTGAPDD340 pKa = 3.15IAVEE344 pKa = 4.02FAEE347 pKa = 4.37YY348 pKa = 10.69LLSDD352 pKa = 3.99EE353 pKa = 4.41VQIGFAEE360 pKa = 4.59LDD362 pKa = 3.24MGLPTNPAATDD373 pKa = 3.44SLANEE378 pKa = 4.35TLAQLIPVRR387 pKa = 11.84DD388 pKa = 4.04GGGKK392 pKa = 6.82TQLYY396 pKa = 10.19LDD398 pKa = 3.84TRR400 pKa = 11.84LGQSVGNALNDD411 pKa = 4.58AIALMFAGQAGPEE424 pKa = 4.48DD425 pKa = 3.47IVEE428 pKa = 4.34ALEE431 pKa = 4.24SASAMGG437 pKa = 3.92
Molecular weight: 45.88 kDa
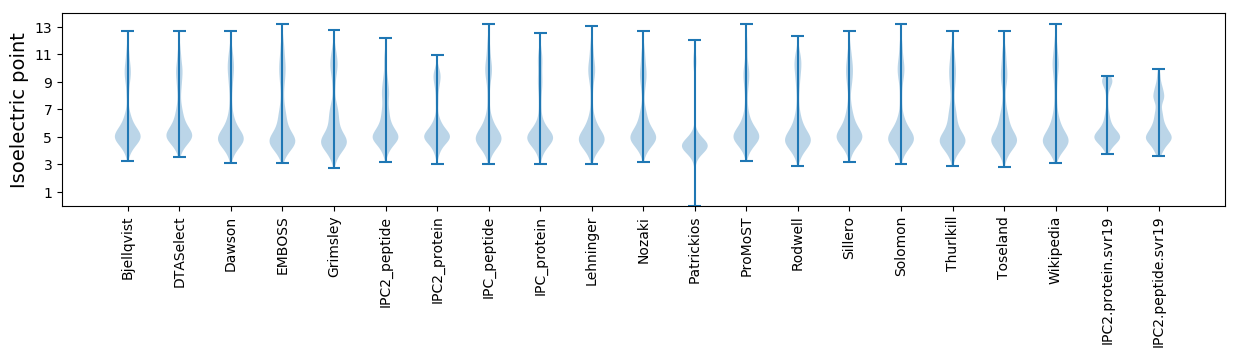
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B9NA99|A0A1B9NA99_9MICO UPF0225 protein A7J15_07480 OS=Microbacterium sediminis OX=904291 GN=A7J15_07480 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.52GRR40 pKa = 11.84TEE42 pKa = 4.14LSAA45 pKa = 4.86
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.52GRR40 pKa = 11.84TEE42 pKa = 4.14LSAA45 pKa = 4.86
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
822206 |
38 |
2094 |
325.1 |
34.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.159 ± 0.09 | 0.477 ± 0.012 |
6.149 ± 0.043 | 6.105 ± 0.047 |
3.032 ± 0.033 | 9.082 ± 0.037 |
1.925 ± 0.023 | 4.696 ± 0.037 |
1.833 ± 0.038 | 10.064 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.83 ± 0.023 | 1.79 ± 0.025 |
5.588 ± 0.036 | 2.73 ± 0.028 |
7.579 ± 0.064 | 4.901 ± 0.031 |
5.762 ± 0.047 | 8.858 ± 0.042 |
1.492 ± 0.022 | 1.949 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
