
Frigoribacterium sp. JB110
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Frigoribacterium; unclassified Frigoribacterium
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
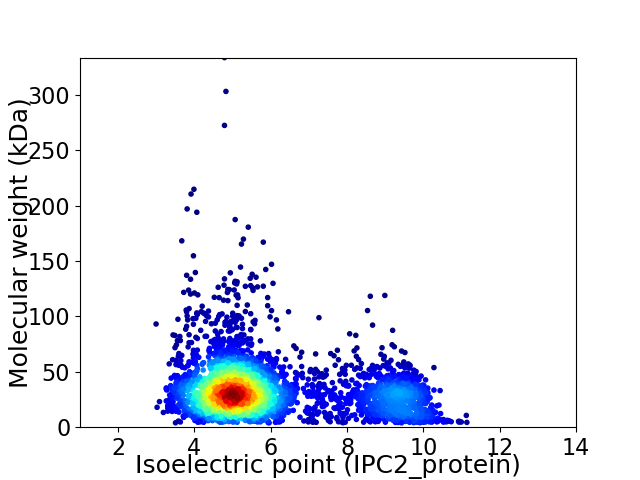
Virtual 2D-PAGE plot for 3430 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1R4GCR7|A0A1R4GCR7_9MICO Uncharacterized protein OS=Frigoribacterium sp. JB110 OX=1434829 GN=CZ774_14100 PE=4 SV=1
MM1 pKa = 7.82RR2 pKa = 11.84AGRR5 pKa = 11.84PKK7 pKa = 10.49RR8 pKa = 11.84RR9 pKa = 11.84IAAAAAVLAAGAMLLSGCLSAFIPEE34 pKa = 4.54RR35 pKa = 11.84PADD38 pKa = 4.03PAPVTDD44 pKa = 4.86GVPSEE49 pKa = 5.2LIEE52 pKa = 6.04FYY54 pKa = 10.64DD55 pKa = 5.87QPPQWQDD62 pKa = 3.17CGPDD66 pKa = 3.72LQCATITAPLDD77 pKa = 3.1WHH79 pKa = 7.07DD80 pKa = 3.87PARR83 pKa = 11.84GEE85 pKa = 4.0IDD87 pKa = 3.53LALTRR92 pKa = 11.84YY93 pKa = 9.3LAEE96 pKa = 3.97QGAPDD101 pKa = 4.78GSLLTNPGGPGASGVSFVQSSAQYY125 pKa = 11.11LFADD129 pKa = 4.65DD130 pKa = 3.96VRR132 pKa = 11.84AAYY135 pKa = 9.98DD136 pKa = 3.15IVGFDD141 pKa = 3.44PRR143 pKa = 11.84GVGEE147 pKa = 4.29STAVEE152 pKa = 4.33CLDD155 pKa = 3.9DD156 pKa = 4.29AAMDD160 pKa = 3.74SYY162 pKa = 11.51LYY164 pKa = 10.15DD165 pKa = 3.71LPEE168 pKa = 4.34GEE170 pKa = 4.52RR171 pKa = 11.84GSAEE175 pKa = 3.79WEE177 pKa = 4.01RR178 pKa = 11.84EE179 pKa = 4.18LEE181 pKa = 4.05KK182 pKa = 11.38SNADD186 pKa = 3.49FAAGCEE192 pKa = 4.27EE193 pKa = 3.99NTGDD197 pKa = 4.14LLEE200 pKa = 5.7FITTEE205 pKa = 3.7QAARR209 pKa = 11.84DD210 pKa = 3.72LDD212 pKa = 4.48LIRR215 pKa = 11.84AVLGEE220 pKa = 4.0NEE222 pKa = 3.75LRR224 pKa = 11.84YY225 pKa = 10.34LGFSYY230 pKa = 9.14GTFLGATYY238 pKa = 11.27ANLFPEE244 pKa = 4.63RR245 pKa = 11.84AGHH248 pKa = 6.48LVLDD252 pKa = 4.28AAIDD256 pKa = 3.93PSVPSAMVGAVQGVGFEE273 pKa = 4.31DD274 pKa = 3.82ALRR277 pKa = 11.84AYY279 pKa = 10.5LADD282 pKa = 4.69CGDD285 pKa = 4.29GGQCPFPGNVDD296 pKa = 4.03DD297 pKa = 6.26GMADD301 pKa = 3.76LGALLAEE308 pKa = 4.54VDD310 pKa = 3.92DD311 pKa = 4.97RR312 pKa = 11.84PLVGADD318 pKa = 3.21GRR320 pKa = 11.84EE321 pKa = 4.01LGADD325 pKa = 3.55TLVTAIISSLYY336 pKa = 10.79SPLSWPDD343 pKa = 2.87LTAALEE349 pKa = 4.48GAQQGDD355 pKa = 3.26ASGAFALADD364 pKa = 3.49MYY366 pKa = 11.42NDD368 pKa = 3.84RR369 pKa = 11.84VNGTYY374 pKa = 9.97VANTTEE380 pKa = 4.62AFSAYY385 pKa = 10.49NCMDD389 pKa = 3.78YY390 pKa = 10.73PVSTAAEE397 pKa = 3.73TDD399 pKa = 3.38AADD402 pKa = 3.81EE403 pKa = 4.37LMSEE407 pKa = 4.4NAPTIAPYY415 pKa = 9.98WSGPDD420 pKa = 3.83LCEE423 pKa = 3.94QWPHH427 pKa = 6.1EE428 pKa = 4.17PTGTRR433 pKa = 11.84DD434 pKa = 3.2RR435 pKa = 11.84ITAEE439 pKa = 3.73GAAPILVVGTTGDD452 pKa = 3.61PATPYY457 pKa = 10.11QWAEE461 pKa = 3.74ALAEE465 pKa = 4.04QLDD468 pKa = 4.38SGTLITFEE476 pKa = 4.6GEE478 pKa = 3.94GHH480 pKa = 6.22GGYY483 pKa = 10.78GNSACINDD491 pKa = 3.72AVGDD495 pKa = 3.83FLIDD499 pKa = 3.26GTVPDD504 pKa = 5.58DD505 pKa = 4.64GLRR508 pKa = 11.84CQQ510 pKa = 4.2
MM1 pKa = 7.82RR2 pKa = 11.84AGRR5 pKa = 11.84PKK7 pKa = 10.49RR8 pKa = 11.84RR9 pKa = 11.84IAAAAAVLAAGAMLLSGCLSAFIPEE34 pKa = 4.54RR35 pKa = 11.84PADD38 pKa = 4.03PAPVTDD44 pKa = 4.86GVPSEE49 pKa = 5.2LIEE52 pKa = 6.04FYY54 pKa = 10.64DD55 pKa = 5.87QPPQWQDD62 pKa = 3.17CGPDD66 pKa = 3.72LQCATITAPLDD77 pKa = 3.1WHH79 pKa = 7.07DD80 pKa = 3.87PARR83 pKa = 11.84GEE85 pKa = 4.0IDD87 pKa = 3.53LALTRR92 pKa = 11.84YY93 pKa = 9.3LAEE96 pKa = 3.97QGAPDD101 pKa = 4.78GSLLTNPGGPGASGVSFVQSSAQYY125 pKa = 11.11LFADD129 pKa = 4.65DD130 pKa = 3.96VRR132 pKa = 11.84AAYY135 pKa = 9.98DD136 pKa = 3.15IVGFDD141 pKa = 3.44PRR143 pKa = 11.84GVGEE147 pKa = 4.29STAVEE152 pKa = 4.33CLDD155 pKa = 3.9DD156 pKa = 4.29AAMDD160 pKa = 3.74SYY162 pKa = 11.51LYY164 pKa = 10.15DD165 pKa = 3.71LPEE168 pKa = 4.34GEE170 pKa = 4.52RR171 pKa = 11.84GSAEE175 pKa = 3.79WEE177 pKa = 4.01RR178 pKa = 11.84EE179 pKa = 4.18LEE181 pKa = 4.05KK182 pKa = 11.38SNADD186 pKa = 3.49FAAGCEE192 pKa = 4.27EE193 pKa = 3.99NTGDD197 pKa = 4.14LLEE200 pKa = 5.7FITTEE205 pKa = 3.7QAARR209 pKa = 11.84DD210 pKa = 3.72LDD212 pKa = 4.48LIRR215 pKa = 11.84AVLGEE220 pKa = 4.0NEE222 pKa = 3.75LRR224 pKa = 11.84YY225 pKa = 10.34LGFSYY230 pKa = 9.14GTFLGATYY238 pKa = 11.27ANLFPEE244 pKa = 4.63RR245 pKa = 11.84AGHH248 pKa = 6.48LVLDD252 pKa = 4.28AAIDD256 pKa = 3.93PSVPSAMVGAVQGVGFEE273 pKa = 4.31DD274 pKa = 3.82ALRR277 pKa = 11.84AYY279 pKa = 10.5LADD282 pKa = 4.69CGDD285 pKa = 4.29GGQCPFPGNVDD296 pKa = 4.03DD297 pKa = 6.26GMADD301 pKa = 3.76LGALLAEE308 pKa = 4.54VDD310 pKa = 3.92DD311 pKa = 4.97RR312 pKa = 11.84PLVGADD318 pKa = 3.21GRR320 pKa = 11.84EE321 pKa = 4.01LGADD325 pKa = 3.55TLVTAIISSLYY336 pKa = 10.79SPLSWPDD343 pKa = 2.87LTAALEE349 pKa = 4.48GAQQGDD355 pKa = 3.26ASGAFALADD364 pKa = 3.49MYY366 pKa = 11.42NDD368 pKa = 3.84RR369 pKa = 11.84VNGTYY374 pKa = 9.97VANTTEE380 pKa = 4.62AFSAYY385 pKa = 10.49NCMDD389 pKa = 3.78YY390 pKa = 10.73PVSTAAEE397 pKa = 3.73TDD399 pKa = 3.38AADD402 pKa = 3.81EE403 pKa = 4.37LMSEE407 pKa = 4.4NAPTIAPYY415 pKa = 9.98WSGPDD420 pKa = 3.83LCEE423 pKa = 3.94QWPHH427 pKa = 6.1EE428 pKa = 4.17PTGTRR433 pKa = 11.84DD434 pKa = 3.2RR435 pKa = 11.84ITAEE439 pKa = 3.73GAAPILVVGTTGDD452 pKa = 3.61PATPYY457 pKa = 10.11QWAEE461 pKa = 3.74ALAEE465 pKa = 4.04QLDD468 pKa = 4.38SGTLITFEE476 pKa = 4.6GEE478 pKa = 3.94GHH480 pKa = 6.22GGYY483 pKa = 10.78GNSACINDD491 pKa = 3.72AVGDD495 pKa = 3.83FLIDD499 pKa = 3.26GTVPDD504 pKa = 5.58DD505 pKa = 4.64GLRR508 pKa = 11.84CQQ510 pKa = 4.2
Molecular weight: 53.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1R4G7I6|A0A1R4G7I6_9MICO Secreted protein OS=Frigoribacterium sp. JB110 OX=1434829 GN=CZ774_12470 PE=4 SV=1
MM1 pKa = 7.29GRR3 pKa = 11.84GAVARR8 pKa = 11.84LGASRR13 pKa = 11.84HH14 pKa = 4.78RR15 pKa = 11.84RR16 pKa = 11.84LIRR19 pKa = 11.84RR20 pKa = 11.84SPARR24 pKa = 11.84KK25 pKa = 9.22ARR27 pKa = 11.84VSGSSLLKK35 pKa = 9.83VTLSLL40 pKa = 4.4
MM1 pKa = 7.29GRR3 pKa = 11.84GAVARR8 pKa = 11.84LGASRR13 pKa = 11.84HH14 pKa = 4.78RR15 pKa = 11.84RR16 pKa = 11.84LIRR19 pKa = 11.84RR20 pKa = 11.84SPARR24 pKa = 11.84KK25 pKa = 9.22ARR27 pKa = 11.84VSGSSLLKK35 pKa = 9.83VTLSLL40 pKa = 4.4
Molecular weight: 4.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1120900 |
37 |
3078 |
326.8 |
35.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.248 ± 0.062 | 0.514 ± 0.011 |
6.635 ± 0.04 | 6.061 ± 0.04 |
3.064 ± 0.025 | 9.066 ± 0.042 |
2.068 ± 0.02 | 4.653 ± 0.027 |
1.823 ± 0.027 | 9.73 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.982 ± 0.018 | 1.949 ± 0.017 |
5.154 ± 0.032 | 2.835 ± 0.023 |
7.354 ± 0.05 | 5.716 ± 0.027 |
6.009 ± 0.034 | 8.783 ± 0.036 |
1.455 ± 0.017 | 1.9 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
