
Aspergillus arachidicola
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Aspergillus
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
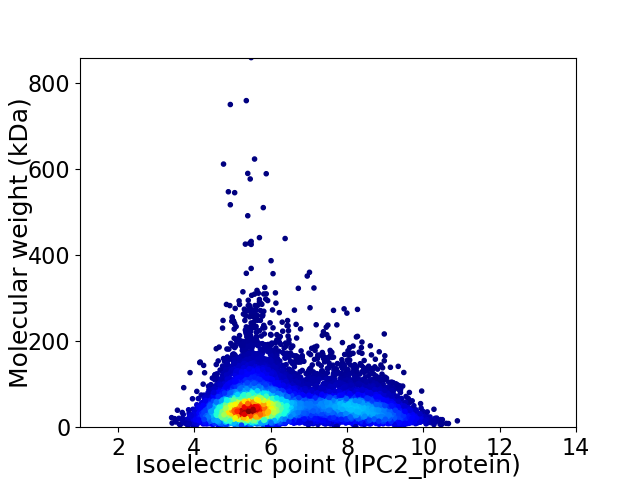
Virtual 2D-PAGE plot for 12090 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2G7EM66|A0A2G7EM66_9EURO Beta-glucosidase (Fragment) OS=Aspergillus arachidicola OX=656916 GN=AARAC_009615 PE=3 SV=1
MM1 pKa = 7.7ADD3 pKa = 3.51EE4 pKa = 5.26AGSIYY9 pKa = 10.71DD10 pKa = 3.85EE11 pKa = 4.95IEE13 pKa = 4.34IEE15 pKa = 5.79DD16 pKa = 3.79MTFDD20 pKa = 6.24PITQLYY26 pKa = 9.73HH27 pKa = 5.63YY28 pKa = 7.55PCPCGDD34 pKa = 3.67RR35 pKa = 11.84FEE37 pKa = 5.7IMIDD41 pKa = 3.49DD42 pKa = 4.19LRR44 pKa = 11.84DD45 pKa = 3.58GEE47 pKa = 4.73EE48 pKa = 3.84IAVCPSCSLKK58 pKa = 10.28IRR60 pKa = 11.84VIFDD64 pKa = 3.02VDD66 pKa = 4.13DD67 pKa = 3.69LHH69 pKa = 9.25KK70 pKa = 10.81DD71 pKa = 3.63DD72 pKa = 4.29QQQGPSAVAVQAA84 pKa = 4.09
MM1 pKa = 7.7ADD3 pKa = 3.51EE4 pKa = 5.26AGSIYY9 pKa = 10.71DD10 pKa = 3.85EE11 pKa = 4.95IEE13 pKa = 4.34IEE15 pKa = 5.79DD16 pKa = 3.79MTFDD20 pKa = 6.24PITQLYY26 pKa = 9.73HH27 pKa = 5.63YY28 pKa = 7.55PCPCGDD34 pKa = 3.67RR35 pKa = 11.84FEE37 pKa = 5.7IMIDD41 pKa = 3.49DD42 pKa = 4.19LRR44 pKa = 11.84DD45 pKa = 3.58GEE47 pKa = 4.73EE48 pKa = 3.84IAVCPSCSLKK58 pKa = 10.28IRR60 pKa = 11.84VIFDD64 pKa = 3.02VDD66 pKa = 4.13DD67 pKa = 3.69LHH69 pKa = 9.25KK70 pKa = 10.81DD71 pKa = 3.63DD72 pKa = 4.29QQQGPSAVAVQAA84 pKa = 4.09
Molecular weight: 9.45 kDa
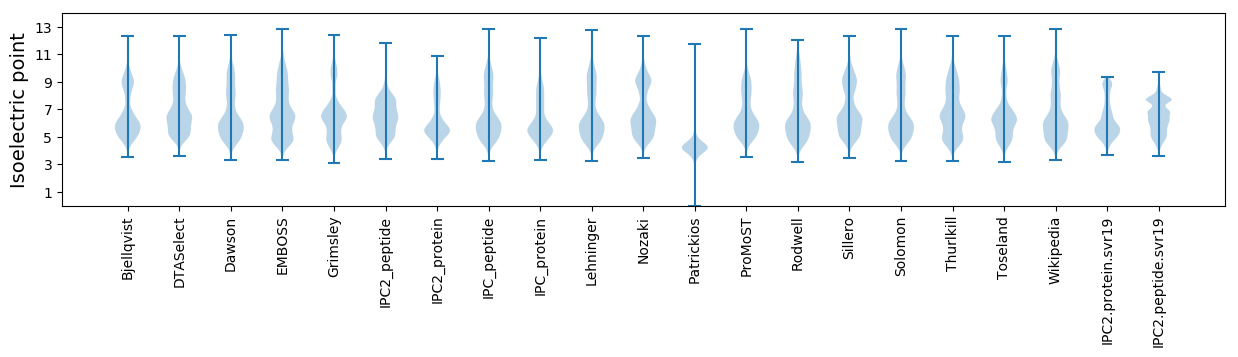
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2G7FUM2|A0A2G7FUM2_9EURO RING-type domain-containing protein OS=Aspergillus arachidicola OX=656916 GN=AARAC_008645 PE=4 SV=1
MM1 pKa = 7.95PNRR4 pKa = 11.84PQFSGRR10 pKa = 11.84TMDD13 pKa = 3.38LTLFSFTRR21 pKa = 11.84AVDD24 pKa = 4.12LVACIIWARR33 pKa = 11.84WRR35 pKa = 11.84RR36 pKa = 11.84WRR38 pKa = 11.84SARR41 pKa = 11.84GRR43 pKa = 11.84WSRR46 pKa = 11.84AEE48 pKa = 3.86SLAPKK53 pKa = 10.04LADD56 pKa = 3.12SGVFAASAAVVMWAWFYY73 pKa = 11.39LPEE76 pKa = 4.93RR77 pKa = 11.84LPKK80 pKa = 10.42SYY82 pKa = 10.54GKK84 pKa = 9.77WIGEE88 pKa = 4.12VAKK91 pKa = 10.63VDD93 pKa = 3.53SRR95 pKa = 11.84LIEE98 pKa = 3.86ALRR101 pKa = 11.84RR102 pKa = 11.84ARR104 pKa = 11.84RR105 pKa = 11.84GVFVYY110 pKa = 10.28GKK112 pKa = 8.77DD113 pKa = 3.14TGQAPLLEE121 pKa = 4.73SMCKK125 pKa = 10.29DD126 pKa = 3.55YY127 pKa = 10.96GWPIEE132 pKa = 4.14WGDD135 pKa = 3.6PSKK138 pKa = 9.97TIPIPCEE145 pKa = 3.88MVHH148 pKa = 6.01MSCGPNCEE156 pKa = 3.69KK157 pKa = 10.72HH158 pKa = 5.65AVSRR162 pKa = 11.84FARR165 pKa = 11.84TFGFACATYY174 pKa = 10.13IPLQIVFRR182 pKa = 11.84LRR184 pKa = 11.84RR185 pKa = 11.84LKK187 pKa = 10.75SVLSLRR193 pKa = 11.84RR194 pKa = 11.84AVSDD198 pKa = 3.34AMRR201 pKa = 11.84SSAFLASFVSIFYY214 pKa = 10.75YY215 pKa = 9.68SVCLARR221 pKa = 11.84TRR223 pKa = 11.84IGPKK227 pKa = 9.14IFPRR231 pKa = 11.84NVVTPMMWDD240 pKa = 2.97SGLCVGAGCLMCGWSILVEE259 pKa = 4.44SPSKK263 pKa = 10.38RR264 pKa = 11.84QEE266 pKa = 3.62LALFVAPRR274 pKa = 11.84AAATVLPRR282 pKa = 11.84FYY284 pKa = 10.89DD285 pKa = 2.98KK286 pKa = 10.71QYY288 pKa = 10.85QYY290 pKa = 11.27RR291 pKa = 11.84EE292 pKa = 4.05RR293 pKa = 11.84IAFAVSAALLLTCLQEE309 pKa = 4.05RR310 pKa = 11.84PGMVRR315 pKa = 11.84GVFGRR320 pKa = 11.84IATSVLKK327 pKa = 10.85
MM1 pKa = 7.95PNRR4 pKa = 11.84PQFSGRR10 pKa = 11.84TMDD13 pKa = 3.38LTLFSFTRR21 pKa = 11.84AVDD24 pKa = 4.12LVACIIWARR33 pKa = 11.84WRR35 pKa = 11.84RR36 pKa = 11.84WRR38 pKa = 11.84SARR41 pKa = 11.84GRR43 pKa = 11.84WSRR46 pKa = 11.84AEE48 pKa = 3.86SLAPKK53 pKa = 10.04LADD56 pKa = 3.12SGVFAASAAVVMWAWFYY73 pKa = 11.39LPEE76 pKa = 4.93RR77 pKa = 11.84LPKK80 pKa = 10.42SYY82 pKa = 10.54GKK84 pKa = 9.77WIGEE88 pKa = 4.12VAKK91 pKa = 10.63VDD93 pKa = 3.53SRR95 pKa = 11.84LIEE98 pKa = 3.86ALRR101 pKa = 11.84RR102 pKa = 11.84ARR104 pKa = 11.84RR105 pKa = 11.84GVFVYY110 pKa = 10.28GKK112 pKa = 8.77DD113 pKa = 3.14TGQAPLLEE121 pKa = 4.73SMCKK125 pKa = 10.29DD126 pKa = 3.55YY127 pKa = 10.96GWPIEE132 pKa = 4.14WGDD135 pKa = 3.6PSKK138 pKa = 9.97TIPIPCEE145 pKa = 3.88MVHH148 pKa = 6.01MSCGPNCEE156 pKa = 3.69KK157 pKa = 10.72HH158 pKa = 5.65AVSRR162 pKa = 11.84FARR165 pKa = 11.84TFGFACATYY174 pKa = 10.13IPLQIVFRR182 pKa = 11.84LRR184 pKa = 11.84RR185 pKa = 11.84LKK187 pKa = 10.75SVLSLRR193 pKa = 11.84RR194 pKa = 11.84AVSDD198 pKa = 3.34AMRR201 pKa = 11.84SSAFLASFVSIFYY214 pKa = 10.75YY215 pKa = 9.68SVCLARR221 pKa = 11.84TRR223 pKa = 11.84IGPKK227 pKa = 9.14IFPRR231 pKa = 11.84NVVTPMMWDD240 pKa = 2.97SGLCVGAGCLMCGWSILVEE259 pKa = 4.44SPSKK263 pKa = 10.38RR264 pKa = 11.84QEE266 pKa = 3.62LALFVAPRR274 pKa = 11.84AAATVLPRR282 pKa = 11.84FYY284 pKa = 10.89DD285 pKa = 2.98KK286 pKa = 10.71QYY288 pKa = 10.85QYY290 pKa = 11.27RR291 pKa = 11.84EE292 pKa = 4.05RR293 pKa = 11.84IAFAVSAALLLTCLQEE309 pKa = 4.05RR310 pKa = 11.84PGMVRR315 pKa = 11.84GVFGRR320 pKa = 11.84IATSVLKK327 pKa = 10.85
Molecular weight: 36.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6134843 |
10 |
7745 |
507.4 |
56.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.269 ± 0.017 | 1.311 ± 0.009 |
5.628 ± 0.014 | 6.101 ± 0.024 |
3.821 ± 0.015 | 6.796 ± 0.02 |
2.448 ± 0.009 | 5.165 ± 0.015 |
4.585 ± 0.018 | 9.225 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.159 ± 0.008 | 3.735 ± 0.009 |
5.836 ± 0.021 | 4.033 ± 0.013 |
5.959 ± 0.019 | 8.225 ± 0.02 |
5.953 ± 0.015 | 6.277 ± 0.015 |
1.526 ± 0.009 | 2.948 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
