
Cyclovirus PKbeef23/PAK/2009
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; Bovine associated cyclovirus 1
Average proteome isoelectric point is 8.42
Get precalculated fractions of proteins
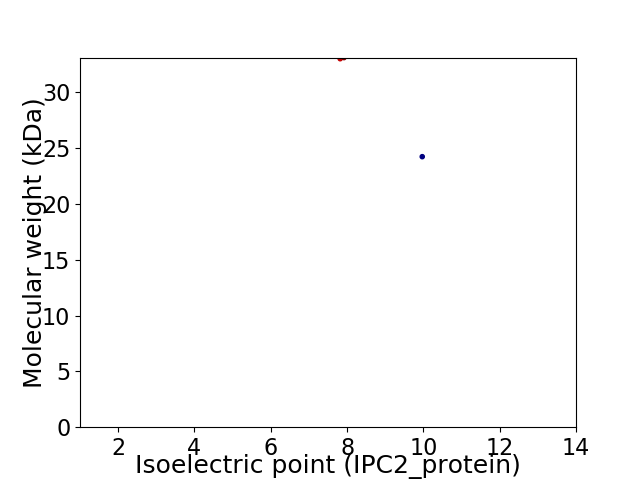
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
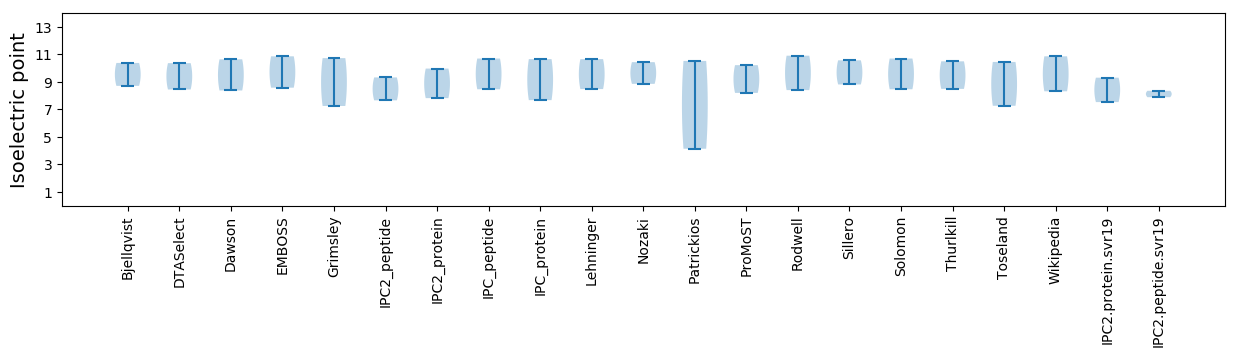
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E9NWS3|E9NWS3_9CIRC Cap protein OS=Cyclovirus PKbeef23/PAK/2009 OX=942032 PE=4 SV=1
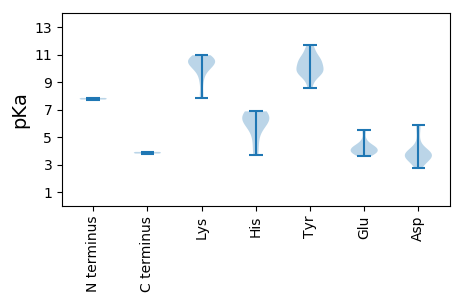
MM1 pKa = 7.72APNKK5 pKa = 7.84TLRR8 pKa = 11.84RR9 pKa = 11.84FCWTLNNYY17 pKa = 8.59TEE19 pKa = 5.48DD20 pKa = 5.87DD21 pKa = 3.54VDD23 pKa = 4.17QLQKK27 pKa = 10.95DD28 pKa = 3.9LPEE31 pKa = 4.39LCKK34 pKa = 9.8FTIFGRR40 pKa = 11.84EE41 pKa = 3.84VCPTTGTKK49 pKa = 9.98HH50 pKa = 6.08LQGFCNLQRR59 pKa = 11.84PKK61 pKa = 10.52RR62 pKa = 11.84FNNIRR67 pKa = 11.84EE68 pKa = 4.13IFKK71 pKa = 10.75GRR73 pKa = 11.84AHH75 pKa = 6.87IEE77 pKa = 3.89GAKK80 pKa = 10.74GSDD83 pKa = 3.17QDD85 pKa = 3.65NQRR88 pKa = 11.84YY89 pKa = 8.69CSKK92 pKa = 10.76SGEE95 pKa = 4.33VWSHH99 pKa = 6.61GEE101 pKa = 3.77PCNQGARR108 pKa = 11.84SDD110 pKa = 3.94LEE112 pKa = 4.17EE113 pKa = 3.9VVSIIKK119 pKa = 10.58GGEE122 pKa = 3.66RR123 pKa = 11.84DD124 pKa = 3.71VKK126 pKa = 10.78AVALQCPTAYY136 pKa = 9.32IKK138 pKa = 10.61YY139 pKa = 9.99FKK141 pKa = 10.74GIEE144 pKa = 3.85NYY146 pKa = 9.58IRR148 pKa = 11.84IYY150 pKa = 10.49HH151 pKa = 6.02ATPEE155 pKa = 4.28RR156 pKa = 11.84DD157 pKa = 3.5FKK159 pKa = 10.88TEE161 pKa = 4.14VYY163 pKa = 9.81FFWGPTGAGKK173 pKa = 10.39SRR175 pKa = 11.84TARR178 pKa = 11.84EE179 pKa = 3.66QALATGLRR187 pKa = 11.84VYY189 pKa = 9.71YY190 pKa = 10.2KK191 pKa = 10.53PRR193 pKa = 11.84GDD195 pKa = 2.71WWDD198 pKa = 3.59GYY200 pKa = 11.08NGHH203 pKa = 6.6EE204 pKa = 4.56CVIFDD209 pKa = 5.69DD210 pKa = 4.91FYY212 pKa = 11.68GWIKK216 pKa = 10.41YY217 pKa = 10.54DD218 pKa = 3.92EE219 pKa = 4.17VLKK222 pKa = 10.89ICDD225 pKa = 3.61RR226 pKa = 11.84YY227 pKa = 9.9PYY229 pKa = 9.86RR230 pKa = 11.84VPVKK234 pKa = 10.28GGYY237 pKa = 9.69EE238 pKa = 3.6NFIAKK243 pKa = 9.92KK244 pKa = 9.87IWFTSNKK251 pKa = 9.17PLEE254 pKa = 4.31QIYY257 pKa = 10.49KK258 pKa = 10.23FIDD261 pKa = 3.61YY262 pKa = 9.7EE263 pKa = 3.92PSAWRR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84LTVEE274 pKa = 4.09RR275 pKa = 11.84EE276 pKa = 3.92FFYY279 pKa = 10.81EE280 pKa = 3.92
MM1 pKa = 7.72APNKK5 pKa = 7.84TLRR8 pKa = 11.84RR9 pKa = 11.84FCWTLNNYY17 pKa = 8.59TEE19 pKa = 5.48DD20 pKa = 5.87DD21 pKa = 3.54VDD23 pKa = 4.17QLQKK27 pKa = 10.95DD28 pKa = 3.9LPEE31 pKa = 4.39LCKK34 pKa = 9.8FTIFGRR40 pKa = 11.84EE41 pKa = 3.84VCPTTGTKK49 pKa = 9.98HH50 pKa = 6.08LQGFCNLQRR59 pKa = 11.84PKK61 pKa = 10.52RR62 pKa = 11.84FNNIRR67 pKa = 11.84EE68 pKa = 4.13IFKK71 pKa = 10.75GRR73 pKa = 11.84AHH75 pKa = 6.87IEE77 pKa = 3.89GAKK80 pKa = 10.74GSDD83 pKa = 3.17QDD85 pKa = 3.65NQRR88 pKa = 11.84YY89 pKa = 8.69CSKK92 pKa = 10.76SGEE95 pKa = 4.33VWSHH99 pKa = 6.61GEE101 pKa = 3.77PCNQGARR108 pKa = 11.84SDD110 pKa = 3.94LEE112 pKa = 4.17EE113 pKa = 3.9VVSIIKK119 pKa = 10.58GGEE122 pKa = 3.66RR123 pKa = 11.84DD124 pKa = 3.71VKK126 pKa = 10.78AVALQCPTAYY136 pKa = 9.32IKK138 pKa = 10.61YY139 pKa = 9.99FKK141 pKa = 10.74GIEE144 pKa = 3.85NYY146 pKa = 9.58IRR148 pKa = 11.84IYY150 pKa = 10.49HH151 pKa = 6.02ATPEE155 pKa = 4.28RR156 pKa = 11.84DD157 pKa = 3.5FKK159 pKa = 10.88TEE161 pKa = 4.14VYY163 pKa = 9.81FFWGPTGAGKK173 pKa = 10.39SRR175 pKa = 11.84TARR178 pKa = 11.84EE179 pKa = 3.66QALATGLRR187 pKa = 11.84VYY189 pKa = 9.71YY190 pKa = 10.2KK191 pKa = 10.53PRR193 pKa = 11.84GDD195 pKa = 2.71WWDD198 pKa = 3.59GYY200 pKa = 11.08NGHH203 pKa = 6.6EE204 pKa = 4.56CVIFDD209 pKa = 5.69DD210 pKa = 4.91FYY212 pKa = 11.68GWIKK216 pKa = 10.41YY217 pKa = 10.54DD218 pKa = 3.92EE219 pKa = 4.17VLKK222 pKa = 10.89ICDD225 pKa = 3.61RR226 pKa = 11.84YY227 pKa = 9.9PYY229 pKa = 9.86RR230 pKa = 11.84VPVKK234 pKa = 10.28GGYY237 pKa = 9.69EE238 pKa = 3.6NFIAKK243 pKa = 9.92KK244 pKa = 9.87IWFTSNKK251 pKa = 9.17PLEE254 pKa = 4.31QIYY257 pKa = 10.49KK258 pKa = 10.23FIDD261 pKa = 3.61YY262 pKa = 9.7EE263 pKa = 3.92PSAWRR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84LTVEE274 pKa = 4.09RR275 pKa = 11.84EE276 pKa = 3.92FFYY279 pKa = 10.81EE280 pKa = 3.92
Molecular weight: 32.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E9NWS3|E9NWS3_9CIRC Cap protein OS=Cyclovirus PKbeef23/PAK/2009 OX=942032 PE=4 SV=1
MM1 pKa = 7.84RR2 pKa = 11.84APIRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.36RR9 pKa = 11.84RR10 pKa = 11.84AIRR13 pKa = 11.84HH14 pKa = 3.72VRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84LRR20 pKa = 11.84KK21 pKa = 7.96VRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84QRR27 pKa = 11.84TGNITVSMKK36 pKa = 10.25SLYY39 pKa = 9.31VAPVEE44 pKa = 4.43GKK46 pKa = 8.28TGYY49 pKa = 10.54LLQISPAVSDD59 pKa = 3.68FTEE62 pKa = 3.83AATFINYY69 pKa = 9.37FEE71 pKa = 5.39AYY73 pKa = 9.75RR74 pKa = 11.84IWSVKK79 pKa = 9.94VKK81 pKa = 10.48VNPLFNVAADD91 pKa = 3.81TTPVPRR97 pKa = 11.84YY98 pKa = 10.02YY99 pKa = 10.49SAPWHH104 pKa = 6.37KK105 pKa = 10.45PGPTSINSTGIVTIDD120 pKa = 2.95KK121 pKa = 10.07CKK123 pKa = 10.6SYY125 pKa = 11.22NGLSGSVRR133 pKa = 11.84RR134 pKa = 11.84FVPAVLTATGYY145 pKa = 11.3AGITTPQYY153 pKa = 11.67GKK155 pKa = 10.43INWRR159 pKa = 11.84PRR161 pKa = 11.84IEE163 pKa = 4.69LNSNTATLPHH173 pKa = 6.27YY174 pKa = 10.42CGLYY178 pKa = 9.91LWSADD183 pKa = 3.5QQEE186 pKa = 4.92GSTHH190 pKa = 4.86IRR192 pKa = 11.84QYY194 pKa = 10.76EE195 pKa = 3.91IEE197 pKa = 4.22TEE199 pKa = 4.16IKK201 pKa = 8.94VTFYY205 pKa = 10.65NQKK208 pKa = 10.49SFLGG212 pKa = 3.8
MM1 pKa = 7.84RR2 pKa = 11.84APIRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.36RR9 pKa = 11.84RR10 pKa = 11.84AIRR13 pKa = 11.84HH14 pKa = 3.72VRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84LRR20 pKa = 11.84KK21 pKa = 7.96VRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84QRR27 pKa = 11.84TGNITVSMKK36 pKa = 10.25SLYY39 pKa = 9.31VAPVEE44 pKa = 4.43GKK46 pKa = 8.28TGYY49 pKa = 10.54LLQISPAVSDD59 pKa = 3.68FTEE62 pKa = 3.83AATFINYY69 pKa = 9.37FEE71 pKa = 5.39AYY73 pKa = 9.75RR74 pKa = 11.84IWSVKK79 pKa = 9.94VKK81 pKa = 10.48VNPLFNVAADD91 pKa = 3.81TTPVPRR97 pKa = 11.84YY98 pKa = 10.02YY99 pKa = 10.49SAPWHH104 pKa = 6.37KK105 pKa = 10.45PGPTSINSTGIVTIDD120 pKa = 2.95KK121 pKa = 10.07CKK123 pKa = 10.6SYY125 pKa = 11.22NGLSGSVRR133 pKa = 11.84RR134 pKa = 11.84FVPAVLTATGYY145 pKa = 11.3AGITTPQYY153 pKa = 11.67GKK155 pKa = 10.43INWRR159 pKa = 11.84PRR161 pKa = 11.84IEE163 pKa = 4.69LNSNTATLPHH173 pKa = 6.27YY174 pKa = 10.42CGLYY178 pKa = 9.91LWSADD183 pKa = 3.5QQEE186 pKa = 4.92GSTHH190 pKa = 4.86IRR192 pKa = 11.84QYY194 pKa = 10.76EE195 pKa = 3.91IEE197 pKa = 4.22TEE199 pKa = 4.16IKK201 pKa = 8.94VTFYY205 pKa = 10.65NQKK208 pKa = 10.49SFLGG212 pKa = 3.8
Molecular weight: 24.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
492 |
212 |
280 |
246.0 |
28.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.894 ± 0.68 | 2.236 ± 0.744 |
4.065 ± 1.254 | 6.098 ± 1.337 |
4.878 ± 0.907 | 7.114 ± 0.565 |
1.829 ± 0.033 | 6.504 ± 0.329 |
6.911 ± 0.719 | 5.285 ± 0.216 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.61 ± 0.192 | 4.472 ± 0.141 |
5.285 ± 0.488 | 3.455 ± 0.088 |
8.74 ± 0.671 | 4.878 ± 1.265 |
6.911 ± 1.181 | 6.098 ± 0.834 |
2.439 ± 0.318 | 6.301 ± 0.097 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
