
Paracoccus denitrificans (strain Pd 1222)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Paracoccus; Paracoccus denitrificans
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
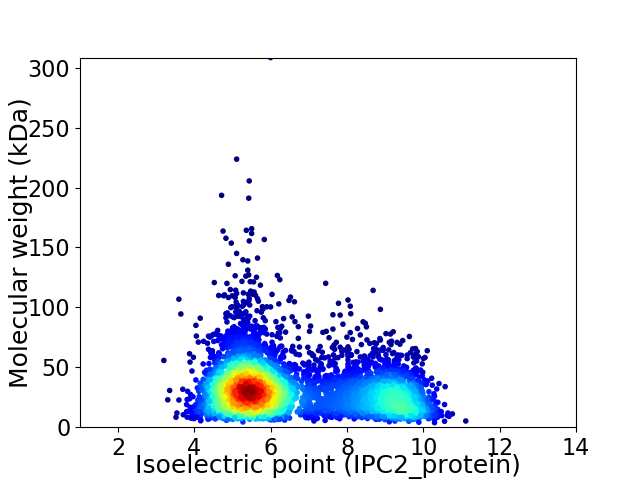
Virtual 2D-PAGE plot for 5019 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
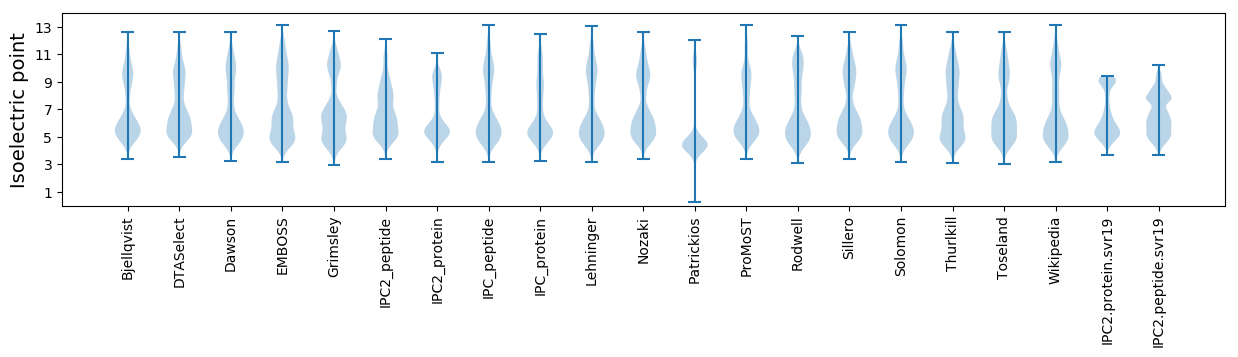
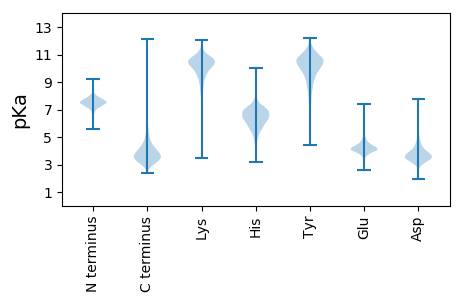
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A1AZI5|A1AZI5_PARDP Succinate dehydrogenase cytochrome b556 subunit OS=Paracoccus denitrificans (strain Pd 1222) OX=318586 GN=Pden_0567 PE=3 SV=1
MM1 pKa = 7.32AQTGRR6 pKa = 11.84ARR8 pKa = 11.84QFLLTSAAGSALFGAHH24 pKa = 5.95ISINLRR30 pKa = 11.84EE31 pKa = 4.2RR32 pKa = 11.84RR33 pKa = 11.84FMATITGTDD42 pKa = 3.16NSEE45 pKa = 4.34TLLGTANADD54 pKa = 4.18LINGLGGNDD63 pKa = 4.11TITGGSGNDD72 pKa = 3.38TMHH75 pKa = 6.9GGSGDD80 pKa = 3.64DD81 pKa = 3.33TFLIGGTDD89 pKa = 3.36IGHH92 pKa = 7.59DD93 pKa = 3.61IYY95 pKa = 11.52DD96 pKa = 4.21GGDD99 pKa = 3.28GADD102 pKa = 3.47QIRR105 pKa = 11.84LNGTLKK111 pKa = 10.73VSNLSLTSANVISTEE126 pKa = 3.83TLNFYY131 pKa = 10.92GYY133 pKa = 10.58GVEE136 pKa = 4.33GTGGNDD142 pKa = 2.92VFDD145 pKa = 4.28ISGMTYY151 pKa = 8.8VSGYY155 pKa = 9.13SRR157 pKa = 11.84IYY159 pKa = 10.12MFDD162 pKa = 3.4GNDD165 pKa = 3.12TFTGYY170 pKa = 10.97AGNDD174 pKa = 3.52NVDD177 pKa = 3.18GGAGNDD183 pKa = 3.73VLNGGAGNDD192 pKa = 3.79TLIGGSGNDD201 pKa = 3.71TLNGGSGNDD210 pKa = 3.25YY211 pKa = 10.73FYY213 pKa = 10.71IGGTNFGHH221 pKa = 7.19DD222 pKa = 3.76VYY224 pKa = 11.49NGGEE228 pKa = 4.09GSDD231 pKa = 4.04EE232 pKa = 3.58IRR234 pKa = 11.84LVGKK238 pKa = 8.02VTVSNLSLTSANVISTEE255 pKa = 3.83TLNFYY260 pKa = 10.92GYY262 pKa = 10.58GVEE265 pKa = 4.33GTGGNDD271 pKa = 2.92VFDD274 pKa = 4.28ISGMTYY280 pKa = 8.8VSGYY284 pKa = 9.13SRR286 pKa = 11.84IYY288 pKa = 10.12MFDD291 pKa = 3.4GNDD294 pKa = 3.12TFTGYY299 pKa = 10.97AGNDD303 pKa = 3.52NVDD306 pKa = 3.18GGAGNDD312 pKa = 3.73VLNGGAGNDD321 pKa = 3.79TLIGGSGNDD330 pKa = 3.71TLNGGSGNDD339 pKa = 3.25YY340 pKa = 10.62FYY342 pKa = 10.74IGGTDD347 pKa = 3.95FGHH350 pKa = 7.14DD351 pKa = 3.51VYY353 pKa = 11.6NGGEE357 pKa = 4.09GSDD360 pKa = 4.04EE361 pKa = 3.58IRR363 pKa = 11.84LVGKK367 pKa = 8.02VTVSNLSLTSANVIGTEE384 pKa = 3.86TLNFYY389 pKa = 10.93GYY391 pKa = 10.58GVEE394 pKa = 4.33GTGGNDD400 pKa = 2.92VFDD403 pKa = 4.28ISGMTYY409 pKa = 8.8VSGYY413 pKa = 9.13SRR415 pKa = 11.84IYY417 pKa = 10.12MFDD420 pKa = 3.4GNDD423 pKa = 3.12TFTGYY428 pKa = 10.97AGNDD432 pKa = 3.52NVDD435 pKa = 3.18GGAGNDD441 pKa = 3.73VLNGGAGNDD450 pKa = 3.79TLIGGSGNDD459 pKa = 3.71TLNGGSGNDD468 pKa = 3.25YY469 pKa = 10.62FYY471 pKa = 10.74IGGTDD476 pKa = 3.95FGHH479 pKa = 7.14DD480 pKa = 3.51VYY482 pKa = 11.6NGGEE486 pKa = 4.09GSDD489 pKa = 4.04EE490 pKa = 3.58IRR492 pKa = 11.84LVGKK496 pKa = 8.02VTVSNLSLTSANVISTEE513 pKa = 3.83TLNFYY518 pKa = 10.9GYY520 pKa = 10.45GIEE523 pKa = 4.3GTGGNDD529 pKa = 2.94VFDD532 pKa = 4.28ISGMTYY538 pKa = 8.8VSGYY542 pKa = 9.56SRR544 pKa = 11.84IYY546 pKa = 9.2MQDD549 pKa = 2.78GNDD552 pKa = 3.35RR553 pKa = 11.84FTGYY557 pKa = 10.85AGNDD561 pKa = 3.52NVDD564 pKa = 3.23GGAGNDD570 pKa = 3.82TLIGGAGNDD579 pKa = 3.87TLNGGSGIDD588 pKa = 3.32TAAYY592 pKa = 9.58GSATAGVTVNLALTTAQAVGGGQGVDD618 pKa = 3.22TLIDD622 pKa = 3.44IEE624 pKa = 4.42NVNGSAYY631 pKa = 10.56ADD633 pKa = 3.42ALLGNAAANLLQGAGGNDD651 pKa = 3.64TLDD654 pKa = 3.68GGAGNDD660 pKa = 3.95TLSGGAGMDD669 pKa = 3.18TAAYY673 pKa = 9.2GRR675 pKa = 11.84ASSGVTVNLGLTGAQLVGGGQGRR698 pKa = 11.84DD699 pKa = 3.29VLRR702 pKa = 11.84SIEE705 pKa = 4.45NIDD708 pKa = 3.28GSGFADD714 pKa = 3.92RR715 pKa = 11.84LTGNAGSNALRR726 pKa = 11.84GGAGNDD732 pKa = 3.51SLSGLGGNDD741 pKa = 2.99RR742 pKa = 11.84LFGGAGADD750 pKa = 3.53QLLGGAGNDD759 pKa = 3.86VLNGDD764 pKa = 4.8AGNDD768 pKa = 3.85LLNGGAGNDD777 pKa = 3.99TLNGGAGIDD786 pKa = 3.4TAAYY790 pKa = 9.06GGASSGVTVNLGLTVAQLIGGGQGRR815 pKa = 11.84DD816 pKa = 3.19VLRR819 pKa = 11.84NIEE822 pKa = 4.13NVNGSGFADD831 pKa = 3.51RR832 pKa = 11.84LTGNTGNNVLNGAAGNDD849 pKa = 3.99TLSGGAGRR857 pKa = 11.84DD858 pKa = 3.56QLLGGAGADD867 pKa = 3.55QLLGGAGNDD876 pKa = 3.89TLNGGAGNDD885 pKa = 3.99TLNGGAGIDD894 pKa = 3.4TAAYY898 pKa = 9.06GGASSGVTVNLGLTVAQLIGGGQGRR923 pKa = 11.84DD924 pKa = 3.19VLRR927 pKa = 11.84NIEE930 pKa = 4.13NVNGSGFADD939 pKa = 3.64RR940 pKa = 11.84LTGNAGNNVLNGAAGNDD957 pKa = 3.99TLSGGAGRR965 pKa = 11.84DD966 pKa = 3.68QLSGGAGADD975 pKa = 3.5RR976 pKa = 11.84LLGGAGNDD984 pKa = 3.89TLNGGAGNDD993 pKa = 3.71TLTGGVGADD1002 pKa = 3.01VFVFAVGNGADD1013 pKa = 5.63RR1014 pKa = 11.84ITDD1017 pKa = 3.48WQDD1020 pKa = 2.79GTDD1023 pKa = 3.63VIRR1026 pKa = 11.84IVSGNWQGEE1035 pKa = 4.51RR1036 pKa = 11.84YY1037 pKa = 10.36DD1038 pKa = 4.22SFDD1041 pKa = 4.54DD1042 pKa = 4.94LSITQSAGNAVVSFGGTTITLAGIGSGQLDD1072 pKa = 3.49ASDD1075 pKa = 4.75FIFVV1079 pKa = 3.67
MM1 pKa = 7.32AQTGRR6 pKa = 11.84ARR8 pKa = 11.84QFLLTSAAGSALFGAHH24 pKa = 5.95ISINLRR30 pKa = 11.84EE31 pKa = 4.2RR32 pKa = 11.84RR33 pKa = 11.84FMATITGTDD42 pKa = 3.16NSEE45 pKa = 4.34TLLGTANADD54 pKa = 4.18LINGLGGNDD63 pKa = 4.11TITGGSGNDD72 pKa = 3.38TMHH75 pKa = 6.9GGSGDD80 pKa = 3.64DD81 pKa = 3.33TFLIGGTDD89 pKa = 3.36IGHH92 pKa = 7.59DD93 pKa = 3.61IYY95 pKa = 11.52DD96 pKa = 4.21GGDD99 pKa = 3.28GADD102 pKa = 3.47QIRR105 pKa = 11.84LNGTLKK111 pKa = 10.73VSNLSLTSANVISTEE126 pKa = 3.83TLNFYY131 pKa = 10.92GYY133 pKa = 10.58GVEE136 pKa = 4.33GTGGNDD142 pKa = 2.92VFDD145 pKa = 4.28ISGMTYY151 pKa = 8.8VSGYY155 pKa = 9.13SRR157 pKa = 11.84IYY159 pKa = 10.12MFDD162 pKa = 3.4GNDD165 pKa = 3.12TFTGYY170 pKa = 10.97AGNDD174 pKa = 3.52NVDD177 pKa = 3.18GGAGNDD183 pKa = 3.73VLNGGAGNDD192 pKa = 3.79TLIGGSGNDD201 pKa = 3.71TLNGGSGNDD210 pKa = 3.25YY211 pKa = 10.73FYY213 pKa = 10.71IGGTNFGHH221 pKa = 7.19DD222 pKa = 3.76VYY224 pKa = 11.49NGGEE228 pKa = 4.09GSDD231 pKa = 4.04EE232 pKa = 3.58IRR234 pKa = 11.84LVGKK238 pKa = 8.02VTVSNLSLTSANVISTEE255 pKa = 3.83TLNFYY260 pKa = 10.92GYY262 pKa = 10.58GVEE265 pKa = 4.33GTGGNDD271 pKa = 2.92VFDD274 pKa = 4.28ISGMTYY280 pKa = 8.8VSGYY284 pKa = 9.13SRR286 pKa = 11.84IYY288 pKa = 10.12MFDD291 pKa = 3.4GNDD294 pKa = 3.12TFTGYY299 pKa = 10.97AGNDD303 pKa = 3.52NVDD306 pKa = 3.18GGAGNDD312 pKa = 3.73VLNGGAGNDD321 pKa = 3.79TLIGGSGNDD330 pKa = 3.71TLNGGSGNDD339 pKa = 3.25YY340 pKa = 10.62FYY342 pKa = 10.74IGGTDD347 pKa = 3.95FGHH350 pKa = 7.14DD351 pKa = 3.51VYY353 pKa = 11.6NGGEE357 pKa = 4.09GSDD360 pKa = 4.04EE361 pKa = 3.58IRR363 pKa = 11.84LVGKK367 pKa = 8.02VTVSNLSLTSANVIGTEE384 pKa = 3.86TLNFYY389 pKa = 10.93GYY391 pKa = 10.58GVEE394 pKa = 4.33GTGGNDD400 pKa = 2.92VFDD403 pKa = 4.28ISGMTYY409 pKa = 8.8VSGYY413 pKa = 9.13SRR415 pKa = 11.84IYY417 pKa = 10.12MFDD420 pKa = 3.4GNDD423 pKa = 3.12TFTGYY428 pKa = 10.97AGNDD432 pKa = 3.52NVDD435 pKa = 3.18GGAGNDD441 pKa = 3.73VLNGGAGNDD450 pKa = 3.79TLIGGSGNDD459 pKa = 3.71TLNGGSGNDD468 pKa = 3.25YY469 pKa = 10.62FYY471 pKa = 10.74IGGTDD476 pKa = 3.95FGHH479 pKa = 7.14DD480 pKa = 3.51VYY482 pKa = 11.6NGGEE486 pKa = 4.09GSDD489 pKa = 4.04EE490 pKa = 3.58IRR492 pKa = 11.84LVGKK496 pKa = 8.02VTVSNLSLTSANVISTEE513 pKa = 3.83TLNFYY518 pKa = 10.9GYY520 pKa = 10.45GIEE523 pKa = 4.3GTGGNDD529 pKa = 2.94VFDD532 pKa = 4.28ISGMTYY538 pKa = 8.8VSGYY542 pKa = 9.56SRR544 pKa = 11.84IYY546 pKa = 9.2MQDD549 pKa = 2.78GNDD552 pKa = 3.35RR553 pKa = 11.84FTGYY557 pKa = 10.85AGNDD561 pKa = 3.52NVDD564 pKa = 3.23GGAGNDD570 pKa = 3.82TLIGGAGNDD579 pKa = 3.87TLNGGSGIDD588 pKa = 3.32TAAYY592 pKa = 9.58GSATAGVTVNLALTTAQAVGGGQGVDD618 pKa = 3.22TLIDD622 pKa = 3.44IEE624 pKa = 4.42NVNGSAYY631 pKa = 10.56ADD633 pKa = 3.42ALLGNAAANLLQGAGGNDD651 pKa = 3.64TLDD654 pKa = 3.68GGAGNDD660 pKa = 3.95TLSGGAGMDD669 pKa = 3.18TAAYY673 pKa = 9.2GRR675 pKa = 11.84ASSGVTVNLGLTGAQLVGGGQGRR698 pKa = 11.84DD699 pKa = 3.29VLRR702 pKa = 11.84SIEE705 pKa = 4.45NIDD708 pKa = 3.28GSGFADD714 pKa = 3.92RR715 pKa = 11.84LTGNAGSNALRR726 pKa = 11.84GGAGNDD732 pKa = 3.51SLSGLGGNDD741 pKa = 2.99RR742 pKa = 11.84LFGGAGADD750 pKa = 3.53QLLGGAGNDD759 pKa = 3.86VLNGDD764 pKa = 4.8AGNDD768 pKa = 3.85LLNGGAGNDD777 pKa = 3.99TLNGGAGIDD786 pKa = 3.4TAAYY790 pKa = 9.06GGASSGVTVNLGLTVAQLIGGGQGRR815 pKa = 11.84DD816 pKa = 3.19VLRR819 pKa = 11.84NIEE822 pKa = 4.13NVNGSGFADD831 pKa = 3.51RR832 pKa = 11.84LTGNTGNNVLNGAAGNDD849 pKa = 3.99TLSGGAGRR857 pKa = 11.84DD858 pKa = 3.56QLLGGAGADD867 pKa = 3.55QLLGGAGNDD876 pKa = 3.89TLNGGAGNDD885 pKa = 3.99TLNGGAGIDD894 pKa = 3.4TAAYY898 pKa = 9.06GGASSGVTVNLGLTVAQLIGGGQGRR923 pKa = 11.84DD924 pKa = 3.19VLRR927 pKa = 11.84NIEE930 pKa = 4.13NVNGSGFADD939 pKa = 3.64RR940 pKa = 11.84LTGNAGNNVLNGAAGNDD957 pKa = 3.99TLSGGAGRR965 pKa = 11.84DD966 pKa = 3.68QLSGGAGADD975 pKa = 3.5RR976 pKa = 11.84LLGGAGNDD984 pKa = 3.89TLNGGAGNDD993 pKa = 3.71TLTGGVGADD1002 pKa = 3.01VFVFAVGNGADD1013 pKa = 5.63RR1014 pKa = 11.84ITDD1017 pKa = 3.48WQDD1020 pKa = 2.79GTDD1023 pKa = 3.63VIRR1026 pKa = 11.84IVSGNWQGEE1035 pKa = 4.51RR1036 pKa = 11.84YY1037 pKa = 10.36DD1038 pKa = 4.22SFDD1041 pKa = 4.54DD1042 pKa = 4.94LSITQSAGNAVVSFGGTTITLAGIGSGQLDD1072 pKa = 3.49ASDD1075 pKa = 4.75FIFVV1079 pKa = 3.67
Molecular weight: 106.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A1B0I1|A1B0I1_PARDP Transcriptional regulator MarR family OS=Paracoccus denitrificans (strain Pd 1222) OX=318586 GN=Pden_0914 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.2QPSNLVRR13 pKa = 11.84ARR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.37GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.13GGRR29 pKa = 11.84RR30 pKa = 11.84VINARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.0GRR40 pKa = 11.84KK41 pKa = 6.51TLSAA45 pKa = 4.15
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.2QPSNLVRR13 pKa = 11.84ARR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.37GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.13GGRR29 pKa = 11.84RR30 pKa = 11.84VINARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.0GRR40 pKa = 11.84KK41 pKa = 6.51TLSAA45 pKa = 4.15
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1561797 |
36 |
2779 |
311.2 |
33.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.254 ± 0.046 | 0.883 ± 0.011 |
5.644 ± 0.028 | 5.71 ± 0.027 |
3.461 ± 0.021 | 9.075 ± 0.037 |
2.1 ± 0.015 | 5.015 ± 0.025 |
2.47 ± 0.026 | 10.442 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.705 ± 0.017 | 2.281 ± 0.019 |
5.601 ± 0.026 | 3.229 ± 0.019 |
7.805 ± 0.035 | 4.854 ± 0.021 |
4.946 ± 0.023 | 7.021 ± 0.026 |
1.475 ± 0.014 | 2.03 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
