
Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Rhodobacter; Rhodobacter capsulatus
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
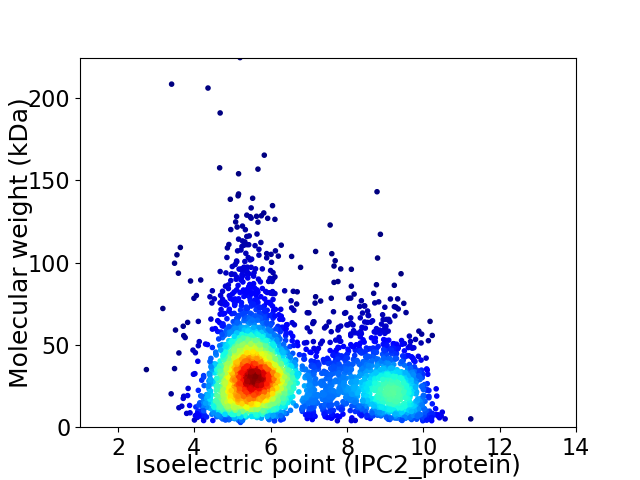
Virtual 2D-PAGE plot for 3632 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
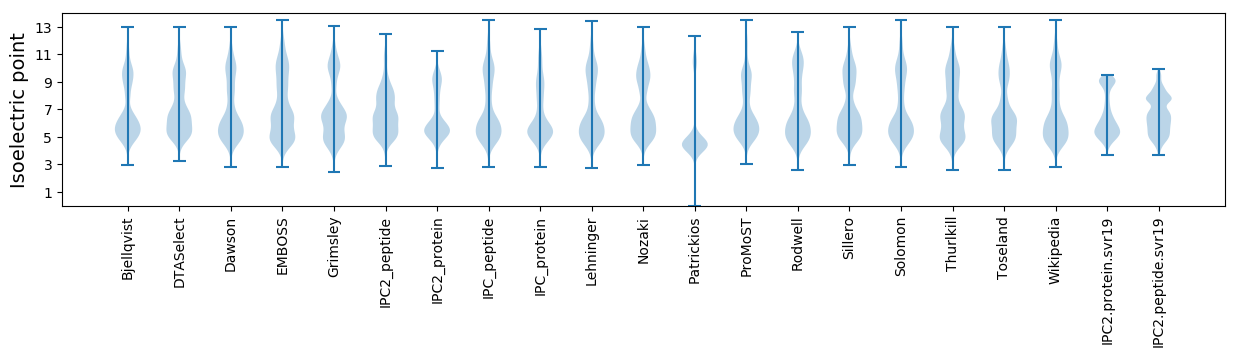
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D5AMD5|D5AMD5_RHOCB zf-CHCC domain-containing protein OS=Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) OX=272942 GN=RCAP_rcc02481 PE=4 SV=1
MM1 pKa = 8.23PITGLPVTSGNNTVTVTPTGIYY23 pKa = 9.84SVNAEE28 pKa = 4.24GGVDD32 pKa = 4.01TLVMDD37 pKa = 4.27YY38 pKa = 10.13STLTSDD44 pKa = 5.23IIYY47 pKa = 9.69EE48 pKa = 4.01YY49 pKa = 10.73AGNGFYY55 pKa = 10.85RR56 pKa = 11.84FTDD59 pKa = 3.81YY60 pKa = 11.1LSSSIDD66 pKa = 3.5FYY68 pKa = 11.49NFEE71 pKa = 4.25RR72 pKa = 11.84FILRR76 pKa = 11.84GGSGDD81 pKa = 4.0DD82 pKa = 4.24DD83 pKa = 3.94LRR85 pKa = 11.84GANDD89 pKa = 2.93VDD91 pKa = 3.68QLYY94 pKa = 10.78GGAGADD100 pKa = 3.68SLTSGLGADD109 pKa = 4.11VVNGGAGFDD118 pKa = 3.37RR119 pKa = 11.84WTAGYY124 pKa = 8.05GTVTGNVLITLSADD138 pKa = 3.32PAITQLVAATGARR151 pKa = 11.84FTQIEE156 pKa = 4.35ALSITTGIGDD166 pKa = 4.0DD167 pKa = 4.48VINTSAFVGNDD178 pKa = 3.94DD179 pKa = 3.57IATGNGSDD187 pKa = 3.47QVNAGRR193 pKa = 11.84GVDD196 pKa = 3.99WINGGEE202 pKa = 4.33GTDD205 pKa = 2.95IMVFDD210 pKa = 4.72WSAITDD216 pKa = 3.58PSFAIRR222 pKa = 11.84NSYY225 pKa = 8.95VSNGWYY231 pKa = 9.89RR232 pKa = 11.84YY233 pKa = 9.21AALSGDD239 pKa = 3.3QVNYY243 pKa = 10.47INFEE247 pKa = 4.27RR248 pKa = 11.84YY249 pKa = 9.13FLTGGAGNDD258 pKa = 3.51WLSGAGQADD267 pKa = 4.37RR268 pKa = 11.84LTGNGGNDD276 pKa = 3.46TLIGGTGADD285 pKa = 4.31TIAGGAGIDD294 pKa = 3.58LWQVDD299 pKa = 4.3LTDD302 pKa = 3.79IEE304 pKa = 5.33ANTSVSLVSQTSNTGAVLSGIEE326 pKa = 3.95RR327 pKa = 11.84LTYY330 pKa = 9.79TGSAFDD336 pKa = 4.14DD337 pKa = 3.77TVVAQSGSYY346 pKa = 10.4NDD348 pKa = 4.08SFTMGAGDD356 pKa = 4.14DD357 pKa = 3.68AVTTGRR363 pKa = 11.84GVDD366 pKa = 3.75GANGEE371 pKa = 4.22AGEE374 pKa = 5.0DD375 pKa = 3.45LLVMNWSGITDD386 pKa = 3.85ATQGISRR393 pKa = 11.84SYY395 pKa = 10.92VSNGWYY401 pKa = 9.94RR402 pKa = 11.84FSAASGDD409 pKa = 3.53RR410 pKa = 11.84LDD412 pKa = 4.38YY413 pKa = 11.3INFEE417 pKa = 4.31RR418 pKa = 11.84FFLTGGAGNDD428 pKa = 3.81YY429 pKa = 11.17LLGAALNDD437 pKa = 3.5RR438 pKa = 11.84LVGNGGDD445 pKa = 3.81DD446 pKa = 3.67TLSGGGGVDD455 pKa = 4.85AITGGDD461 pKa = 4.43GVDD464 pKa = 2.93LWEE467 pKa = 5.09ANLSEE472 pKa = 5.07HH473 pKa = 5.66EE474 pKa = 4.47TNVTINFQTQTTNYY488 pKa = 8.56GAVISGLEE496 pKa = 3.86RR497 pKa = 11.84LNYY500 pKa = 9.57TGTAVRR506 pKa = 11.84DD507 pKa = 4.24LITTRR512 pKa = 11.84SGAYY516 pKa = 9.85DD517 pKa = 3.61DD518 pKa = 4.81NLYY521 pKa = 10.89LGAGNDD527 pKa = 3.54SAAVWRR533 pKa = 11.84GVDD536 pKa = 3.33AVNGAEE542 pKa = 5.07GEE544 pKa = 4.27DD545 pKa = 3.82TLILDD550 pKa = 4.01WSGIAGATQGITYY563 pKa = 10.01SYY565 pKa = 10.85VSNGWYY571 pKa = 10.01RR572 pKa = 11.84FSSTSGDD579 pKa = 3.23QVDD582 pKa = 5.49FINIEE587 pKa = 4.2HH588 pKa = 6.87YY589 pKa = 11.51DD590 pKa = 3.5MTGGAGHH597 pKa = 7.56DD598 pKa = 3.86SLVGGTMNDD607 pKa = 3.59TLRR610 pKa = 11.84GGAGNDD616 pKa = 3.7TLDD619 pKa = 3.7SGAGRR624 pKa = 11.84AVIDD628 pKa = 4.03GGAGTDD634 pKa = 3.16LWRR637 pKa = 11.84ADD639 pKa = 3.91LSAISAGLRR648 pKa = 11.84FSASASQTAAQATGSGCDD666 pKa = 3.23VRR668 pKa = 11.84NIEE671 pKa = 4.3QVSLTGGAGADD682 pKa = 4.32LIDD685 pKa = 3.67TTGQSGNDD693 pKa = 3.2WFAGNAGNDD702 pKa = 3.81TVSLGLGQDD711 pKa = 3.74GFDD714 pKa = 3.7GGAGVDD720 pKa = 4.96LLILDD725 pKa = 4.4YY726 pKa = 11.01SASTEE731 pKa = 4.45AIQSRR736 pKa = 11.84YY737 pKa = 7.43TSNGWNRR744 pKa = 11.84YY745 pKa = 8.07GEE747 pKa = 4.02QGDD750 pKa = 3.87AHH752 pKa = 6.66FLDD755 pKa = 4.81YY756 pKa = 11.33INVEE760 pKa = 4.17RR761 pKa = 11.84FNVTGGAGADD771 pKa = 3.57TLIGAGAADD780 pKa = 4.52TLTGGAGNDD789 pKa = 3.6WLNGGAGADD798 pKa = 4.11VINGGAGVDD807 pKa = 3.19TWQASFAGATVAIGLTLSAAGAATVSGAGTVLTGIEE843 pKa = 4.3AVSLTTGQNADD854 pKa = 3.6IVNLSAGTGNDD865 pKa = 2.79AVATNEE871 pKa = 4.16GNDD874 pKa = 3.65TVNLGRR880 pKa = 11.84GRR882 pKa = 11.84SEE884 pKa = 3.87WADD887 pKa = 3.16GGAGTDD893 pKa = 3.54SLTLNASLATSGLRR907 pKa = 11.84MDD909 pKa = 4.15YY910 pKa = 10.77VSNGAYY916 pKa = 10.01HH917 pKa = 7.57IYY919 pKa = 9.57ATDD922 pKa = 3.59GSYY925 pKa = 11.38DD926 pKa = 3.17ATFNNFEE933 pKa = 4.53TYY935 pKa = 10.62NITGSARR942 pKa = 11.84NDD944 pKa = 2.98RR945 pKa = 11.84LYY947 pKa = 11.31AFGGNDD953 pKa = 4.19RR954 pKa = 11.84ISTGAGNDD962 pKa = 3.23ILNGGDD968 pKa = 4.25GNDD971 pKa = 3.8TLLGGAGADD980 pKa = 3.07VFQFTNLYY988 pKa = 9.91GCGIDD993 pKa = 4.18TIADD997 pKa = 3.66GAIGDD1002 pKa = 4.09RR1003 pKa = 11.84LRR1005 pKa = 11.84LSGLALSSIANGAGTTLGAGQIAVSASGGVTTLAIGLDD1043 pKa = 3.83GTAGADD1049 pKa = 3.22LTIRR1053 pKa = 11.84LTGSFTAADD1062 pKa = 4.51FSLSGSDD1069 pKa = 3.92VIFII1073 pKa = 4.75
MM1 pKa = 8.23PITGLPVTSGNNTVTVTPTGIYY23 pKa = 9.84SVNAEE28 pKa = 4.24GGVDD32 pKa = 4.01TLVMDD37 pKa = 4.27YY38 pKa = 10.13STLTSDD44 pKa = 5.23IIYY47 pKa = 9.69EE48 pKa = 4.01YY49 pKa = 10.73AGNGFYY55 pKa = 10.85RR56 pKa = 11.84FTDD59 pKa = 3.81YY60 pKa = 11.1LSSSIDD66 pKa = 3.5FYY68 pKa = 11.49NFEE71 pKa = 4.25RR72 pKa = 11.84FILRR76 pKa = 11.84GGSGDD81 pKa = 4.0DD82 pKa = 4.24DD83 pKa = 3.94LRR85 pKa = 11.84GANDD89 pKa = 2.93VDD91 pKa = 3.68QLYY94 pKa = 10.78GGAGADD100 pKa = 3.68SLTSGLGADD109 pKa = 4.11VVNGGAGFDD118 pKa = 3.37RR119 pKa = 11.84WTAGYY124 pKa = 8.05GTVTGNVLITLSADD138 pKa = 3.32PAITQLVAATGARR151 pKa = 11.84FTQIEE156 pKa = 4.35ALSITTGIGDD166 pKa = 4.0DD167 pKa = 4.48VINTSAFVGNDD178 pKa = 3.94DD179 pKa = 3.57IATGNGSDD187 pKa = 3.47QVNAGRR193 pKa = 11.84GVDD196 pKa = 3.99WINGGEE202 pKa = 4.33GTDD205 pKa = 2.95IMVFDD210 pKa = 4.72WSAITDD216 pKa = 3.58PSFAIRR222 pKa = 11.84NSYY225 pKa = 8.95VSNGWYY231 pKa = 9.89RR232 pKa = 11.84YY233 pKa = 9.21AALSGDD239 pKa = 3.3QVNYY243 pKa = 10.47INFEE247 pKa = 4.27RR248 pKa = 11.84YY249 pKa = 9.13FLTGGAGNDD258 pKa = 3.51WLSGAGQADD267 pKa = 4.37RR268 pKa = 11.84LTGNGGNDD276 pKa = 3.46TLIGGTGADD285 pKa = 4.31TIAGGAGIDD294 pKa = 3.58LWQVDD299 pKa = 4.3LTDD302 pKa = 3.79IEE304 pKa = 5.33ANTSVSLVSQTSNTGAVLSGIEE326 pKa = 3.95RR327 pKa = 11.84LTYY330 pKa = 9.79TGSAFDD336 pKa = 4.14DD337 pKa = 3.77TVVAQSGSYY346 pKa = 10.4NDD348 pKa = 4.08SFTMGAGDD356 pKa = 4.14DD357 pKa = 3.68AVTTGRR363 pKa = 11.84GVDD366 pKa = 3.75GANGEE371 pKa = 4.22AGEE374 pKa = 5.0DD375 pKa = 3.45LLVMNWSGITDD386 pKa = 3.85ATQGISRR393 pKa = 11.84SYY395 pKa = 10.92VSNGWYY401 pKa = 9.94RR402 pKa = 11.84FSAASGDD409 pKa = 3.53RR410 pKa = 11.84LDD412 pKa = 4.38YY413 pKa = 11.3INFEE417 pKa = 4.31RR418 pKa = 11.84FFLTGGAGNDD428 pKa = 3.81YY429 pKa = 11.17LLGAALNDD437 pKa = 3.5RR438 pKa = 11.84LVGNGGDD445 pKa = 3.81DD446 pKa = 3.67TLSGGGGVDD455 pKa = 4.85AITGGDD461 pKa = 4.43GVDD464 pKa = 2.93LWEE467 pKa = 5.09ANLSEE472 pKa = 5.07HH473 pKa = 5.66EE474 pKa = 4.47TNVTINFQTQTTNYY488 pKa = 8.56GAVISGLEE496 pKa = 3.86RR497 pKa = 11.84LNYY500 pKa = 9.57TGTAVRR506 pKa = 11.84DD507 pKa = 4.24LITTRR512 pKa = 11.84SGAYY516 pKa = 9.85DD517 pKa = 3.61DD518 pKa = 4.81NLYY521 pKa = 10.89LGAGNDD527 pKa = 3.54SAAVWRR533 pKa = 11.84GVDD536 pKa = 3.33AVNGAEE542 pKa = 5.07GEE544 pKa = 4.27DD545 pKa = 3.82TLILDD550 pKa = 4.01WSGIAGATQGITYY563 pKa = 10.01SYY565 pKa = 10.85VSNGWYY571 pKa = 10.01RR572 pKa = 11.84FSSTSGDD579 pKa = 3.23QVDD582 pKa = 5.49FINIEE587 pKa = 4.2HH588 pKa = 6.87YY589 pKa = 11.51DD590 pKa = 3.5MTGGAGHH597 pKa = 7.56DD598 pKa = 3.86SLVGGTMNDD607 pKa = 3.59TLRR610 pKa = 11.84GGAGNDD616 pKa = 3.7TLDD619 pKa = 3.7SGAGRR624 pKa = 11.84AVIDD628 pKa = 4.03GGAGTDD634 pKa = 3.16LWRR637 pKa = 11.84ADD639 pKa = 3.91LSAISAGLRR648 pKa = 11.84FSASASQTAAQATGSGCDD666 pKa = 3.23VRR668 pKa = 11.84NIEE671 pKa = 4.3QVSLTGGAGADD682 pKa = 4.32LIDD685 pKa = 3.67TTGQSGNDD693 pKa = 3.2WFAGNAGNDD702 pKa = 3.81TVSLGLGQDD711 pKa = 3.74GFDD714 pKa = 3.7GGAGVDD720 pKa = 4.96LLILDD725 pKa = 4.4YY726 pKa = 11.01SASTEE731 pKa = 4.45AIQSRR736 pKa = 11.84YY737 pKa = 7.43TSNGWNRR744 pKa = 11.84YY745 pKa = 8.07GEE747 pKa = 4.02QGDD750 pKa = 3.87AHH752 pKa = 6.66FLDD755 pKa = 4.81YY756 pKa = 11.33INVEE760 pKa = 4.17RR761 pKa = 11.84FNVTGGAGADD771 pKa = 3.57TLIGAGAADD780 pKa = 4.52TLTGGAGNDD789 pKa = 3.6WLNGGAGADD798 pKa = 4.11VINGGAGVDD807 pKa = 3.19TWQASFAGATVAIGLTLSAAGAATVSGAGTVLTGIEE843 pKa = 4.3AVSLTTGQNADD854 pKa = 3.6IVNLSAGTGNDD865 pKa = 2.79AVATNEE871 pKa = 4.16GNDD874 pKa = 3.65TVNLGRR880 pKa = 11.84GRR882 pKa = 11.84SEE884 pKa = 3.87WADD887 pKa = 3.16GGAGTDD893 pKa = 3.54SLTLNASLATSGLRR907 pKa = 11.84MDD909 pKa = 4.15YY910 pKa = 10.77VSNGAYY916 pKa = 10.01HH917 pKa = 7.57IYY919 pKa = 9.57ATDD922 pKa = 3.59GSYY925 pKa = 11.38DD926 pKa = 3.17ATFNNFEE933 pKa = 4.53TYY935 pKa = 10.62NITGSARR942 pKa = 11.84NDD944 pKa = 2.98RR945 pKa = 11.84LYY947 pKa = 11.31AFGGNDD953 pKa = 4.19RR954 pKa = 11.84ISTGAGNDD962 pKa = 3.23ILNGGDD968 pKa = 4.25GNDD971 pKa = 3.8TLLGGAGADD980 pKa = 3.07VFQFTNLYY988 pKa = 9.91GCGIDD993 pKa = 4.18TIADD997 pKa = 3.66GAIGDD1002 pKa = 4.09RR1003 pKa = 11.84LRR1005 pKa = 11.84LSGLALSSIANGAGTTLGAGQIAVSASGGVTTLAIGLDD1043 pKa = 3.83GTAGADD1049 pKa = 3.22LTIRR1053 pKa = 11.84LTGSFTAADD1062 pKa = 4.51FSLSGSDD1069 pKa = 3.92VIFII1073 pKa = 4.75
Molecular weight: 109.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D5AM50|D5AM50_RHOCB S1 RNA binding domain protein OS=Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) OX=272942 GN=RCAP_rcc00355 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.43GGRR28 pKa = 11.84KK29 pKa = 8.99VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97VLSAA44 pKa = 4.11
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.43GGRR28 pKa = 11.84KK29 pKa = 8.99VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97VLSAA44 pKa = 4.11
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1160234 |
28 |
2145 |
319.4 |
34.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.118 ± 0.079 | 0.878 ± 0.013 |
5.491 ± 0.037 | 5.738 ± 0.044 |
3.567 ± 0.025 | 8.913 ± 0.047 |
1.922 ± 0.018 | 4.742 ± 0.03 |
2.983 ± 0.037 | 10.712 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.558 ± 0.022 | 2.12 ± 0.025 |
5.553 ± 0.041 | 3.025 ± 0.024 |
7.241 ± 0.044 | 4.648 ± 0.029 |
5.39 ± 0.037 | 7.102 ± 0.035 |
1.367 ± 0.017 | 1.932 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
