
Synechococcus phage S-RIM8
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Neptunevirus; Synechococcus virus SRIM8
Average proteome isoelectric point is 5.74
Get precalculated fractions of proteins
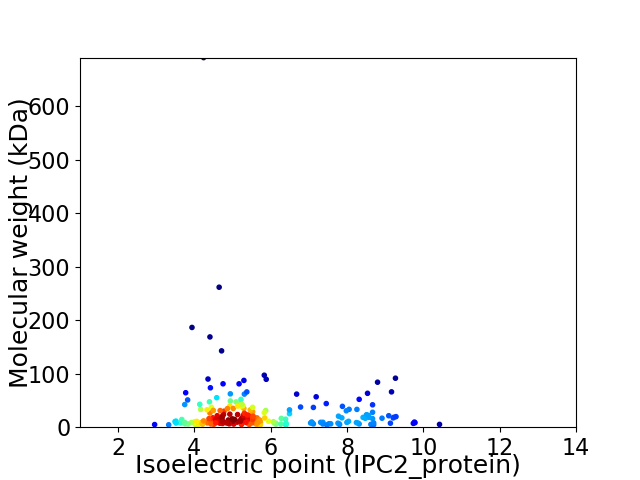
Virtual 2D-PAGE plot for 222 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
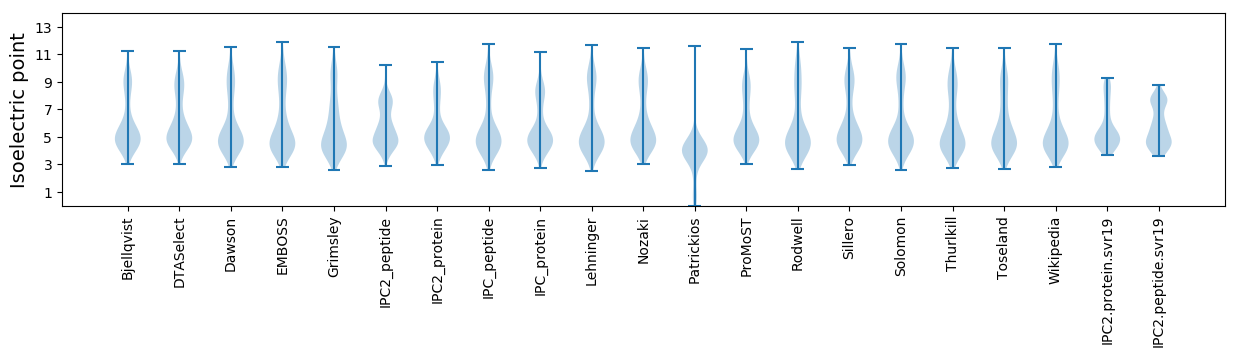
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D7SAI4|A0A1D7SAI4_9CAUD Uncharacterized protein OS=Synechococcus phage S-RIM8 OX=756278 GN=RW010115_044 PE=4 SV=1
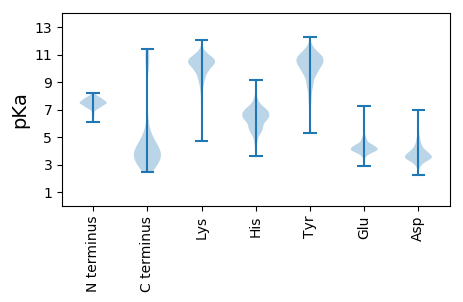
MM1 pKa = 7.54ARR3 pKa = 11.84QGLLAQSKK11 pKa = 9.38PLAATDD17 pKa = 3.58TLLYY21 pKa = 9.81SAPVDD26 pKa = 3.79TSASAVLKK34 pKa = 10.2IANDD38 pKa = 4.23GTGSAYY44 pKa = 10.53SVALRR49 pKa = 11.84DD50 pKa = 3.63YY51 pKa = 11.23DD52 pKa = 3.43QDD54 pKa = 3.93LVLDD58 pKa = 3.83SSSYY62 pKa = 10.93LLHH65 pKa = 7.13KK66 pKa = 10.61GDD68 pKa = 3.53IVTSYY73 pKa = 10.54RR74 pKa = 11.84VSIDD78 pKa = 3.17TAVSVGSFTPGAQFVSSSGEE98 pKa = 3.77STMKK102 pKa = 10.59FEE104 pKa = 4.59SFYY107 pKa = 11.21VPPLTTIYY115 pKa = 11.13VKK117 pKa = 8.26TTSIRR122 pKa = 11.84VISLEE127 pKa = 4.21LVDD130 pKa = 3.83TATGEE135 pKa = 4.26FNVGDD140 pKa = 4.03TVSKK144 pKa = 10.86GVAPDD149 pKa = 3.27NTDD152 pKa = 2.93AVIYY156 pKa = 9.93DD157 pKa = 3.63VNGDD161 pKa = 4.81FITIGPSTINGLGTEE176 pKa = 4.66FVDD179 pKa = 4.28GDD181 pKa = 4.89AITSSSTASGTIAAGGISATEE202 pKa = 3.65NKK204 pKa = 10.21FIFSEE209 pKa = 4.53TTVGGTYY216 pKa = 10.39SRR218 pKa = 11.84TVSDD222 pKa = 4.18LFSDD226 pKa = 3.9RR227 pKa = 11.84VYY229 pKa = 11.0RR230 pKa = 11.84FDD232 pKa = 4.6VSDD235 pKa = 3.28TSMTGLLFALSEE247 pKa = 4.45TEE249 pKa = 3.94NGEE252 pKa = 3.97WGPDD256 pKa = 3.07GLAPTDD262 pKa = 4.39PNDD265 pKa = 3.95ALDD268 pKa = 4.1GGIEE272 pKa = 4.0YY273 pKa = 9.25TIGKK277 pKa = 5.34TTNGTAGSGGAYY289 pKa = 8.99VQYY292 pKa = 11.58DD293 pKa = 3.56MAANTSLPTSLFWYY307 pKa = 7.33EE308 pKa = 3.77TDD310 pKa = 3.37LTTAANASYY319 pKa = 11.07GGGQDD324 pKa = 3.28SFSTSSEE331 pKa = 3.75VTYY334 pKa = 10.79TEE336 pKa = 5.39FYY338 pKa = 10.69AYY340 pKa = 10.37DD341 pKa = 4.68LDD343 pKa = 4.38GTWTNNVDD351 pKa = 3.45SFEE354 pKa = 5.75DD355 pKa = 3.44NFITYY360 pKa = 7.85TVEE363 pKa = 4.1GQTSGPYY370 pKa = 10.25GVVSDD375 pKa = 4.03YY376 pKa = 11.31TGTSLKK382 pKa = 10.3IILGVGSAEE391 pKa = 3.94FAGSNTFFDD400 pKa = 4.02VPKK403 pKa = 11.04SNTADD408 pKa = 3.87RR409 pKa = 11.84NTVTVSSVTTAKK421 pKa = 9.57STIAADD427 pKa = 3.59SYY429 pKa = 11.69LVDD432 pKa = 4.44GNTNSANNTDD442 pKa = 3.79TVTSLVVGPGQRR454 pKa = 11.84VHH456 pKa = 6.54VNSVTANNVFSLIGFEE472 pKa = 4.94DD473 pKa = 4.66GSSEE477 pKa = 4.1YY478 pKa = 7.39TTRR481 pKa = 11.84VYY483 pKa = 11.13GQTT486 pKa = 3.3
MM1 pKa = 7.54ARR3 pKa = 11.84QGLLAQSKK11 pKa = 9.38PLAATDD17 pKa = 3.58TLLYY21 pKa = 9.81SAPVDD26 pKa = 3.79TSASAVLKK34 pKa = 10.2IANDD38 pKa = 4.23GTGSAYY44 pKa = 10.53SVALRR49 pKa = 11.84DD50 pKa = 3.63YY51 pKa = 11.23DD52 pKa = 3.43QDD54 pKa = 3.93LVLDD58 pKa = 3.83SSSYY62 pKa = 10.93LLHH65 pKa = 7.13KK66 pKa = 10.61GDD68 pKa = 3.53IVTSYY73 pKa = 10.54RR74 pKa = 11.84VSIDD78 pKa = 3.17TAVSVGSFTPGAQFVSSSGEE98 pKa = 3.77STMKK102 pKa = 10.59FEE104 pKa = 4.59SFYY107 pKa = 11.21VPPLTTIYY115 pKa = 11.13VKK117 pKa = 8.26TTSIRR122 pKa = 11.84VISLEE127 pKa = 4.21LVDD130 pKa = 3.83TATGEE135 pKa = 4.26FNVGDD140 pKa = 4.03TVSKK144 pKa = 10.86GVAPDD149 pKa = 3.27NTDD152 pKa = 2.93AVIYY156 pKa = 9.93DD157 pKa = 3.63VNGDD161 pKa = 4.81FITIGPSTINGLGTEE176 pKa = 4.66FVDD179 pKa = 4.28GDD181 pKa = 4.89AITSSSTASGTIAAGGISATEE202 pKa = 3.65NKK204 pKa = 10.21FIFSEE209 pKa = 4.53TTVGGTYY216 pKa = 10.39SRR218 pKa = 11.84TVSDD222 pKa = 4.18LFSDD226 pKa = 3.9RR227 pKa = 11.84VYY229 pKa = 11.0RR230 pKa = 11.84FDD232 pKa = 4.6VSDD235 pKa = 3.28TSMTGLLFALSEE247 pKa = 4.45TEE249 pKa = 3.94NGEE252 pKa = 3.97WGPDD256 pKa = 3.07GLAPTDD262 pKa = 4.39PNDD265 pKa = 3.95ALDD268 pKa = 4.1GGIEE272 pKa = 4.0YY273 pKa = 9.25TIGKK277 pKa = 5.34TTNGTAGSGGAYY289 pKa = 8.99VQYY292 pKa = 11.58DD293 pKa = 3.56MAANTSLPTSLFWYY307 pKa = 7.33EE308 pKa = 3.77TDD310 pKa = 3.37LTTAANASYY319 pKa = 11.07GGGQDD324 pKa = 3.28SFSTSSEE331 pKa = 3.75VTYY334 pKa = 10.79TEE336 pKa = 5.39FYY338 pKa = 10.69AYY340 pKa = 10.37DD341 pKa = 4.68LDD343 pKa = 4.38GTWTNNVDD351 pKa = 3.45SFEE354 pKa = 5.75DD355 pKa = 3.44NFITYY360 pKa = 7.85TVEE363 pKa = 4.1GQTSGPYY370 pKa = 10.25GVVSDD375 pKa = 4.03YY376 pKa = 11.31TGTSLKK382 pKa = 10.3IILGVGSAEE391 pKa = 3.94FAGSNTFFDD400 pKa = 4.02VPKK403 pKa = 11.04SNTADD408 pKa = 3.87RR409 pKa = 11.84NTVTVSSVTTAKK421 pKa = 9.57STIAADD427 pKa = 3.59SYY429 pKa = 11.69LVDD432 pKa = 4.44GNTNSANNTDD442 pKa = 3.79TVTSLVVGPGQRR454 pKa = 11.84VHH456 pKa = 6.54VNSVTANNVFSLIGFEE472 pKa = 4.94DD473 pKa = 4.66GSSEE477 pKa = 4.1YY478 pKa = 7.39TTRR481 pKa = 11.84VYY483 pKa = 11.13GQTT486 pKa = 3.3
Molecular weight: 50.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D7SCL2|A0A1D7SCL2_9CAUD Uncharacterized protein OS=Synechococcus phage S-RIM8 OX=756278 GN=RW010115_102 PE=4 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84ALTKK6 pKa = 9.73TRR8 pKa = 11.84SAKK11 pKa = 10.25FYY13 pKa = 9.61SNQVKK18 pKa = 9.8FLLLIVIAVLFWNSNDD34 pKa = 2.83ARR36 pKa = 11.84RR37 pKa = 11.84FTSDD41 pKa = 3.17RR42 pKa = 11.84LNDD45 pKa = 3.33AAEE48 pKa = 4.35FVRR51 pKa = 11.84PANNEE56 pKa = 3.21IRR58 pKa = 11.84ISFF61 pKa = 3.8
MM1 pKa = 7.86RR2 pKa = 11.84ALTKK6 pKa = 9.73TRR8 pKa = 11.84SAKK11 pKa = 10.25FYY13 pKa = 9.61SNQVKK18 pKa = 9.8FLLLIVIAVLFWNSNDD34 pKa = 2.83ARR36 pKa = 11.84RR37 pKa = 11.84FTSDD41 pKa = 3.17RR42 pKa = 11.84LNDD45 pKa = 3.33AAEE48 pKa = 4.35FVRR51 pKa = 11.84PANNEE56 pKa = 3.21IRR58 pKa = 11.84ISFF61 pKa = 3.8
Molecular weight: 7.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
54758 |
37 |
6395 |
246.7 |
27.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.042 ± 0.2 | 0.882 ± 0.093 |
6.616 ± 0.148 | 6.331 ± 0.28 |
4.304 ± 0.122 | 7.639 ± 0.304 |
1.443 ± 0.132 | 6.538 ± 0.214 |
5.711 ± 0.391 | 7.277 ± 0.159 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.987 ± 0.219 | 5.785 ± 0.205 |
3.861 ± 0.13 | 3.875 ± 0.084 |
4.124 ± 0.168 | 6.967 ± 0.224 |
7.349 ± 0.46 | 6.846 ± 0.201 |
1.123 ± 0.098 | 4.299 ± 0.143 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
