
Salmovirus WFRC1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.31
Get precalculated fractions of proteins
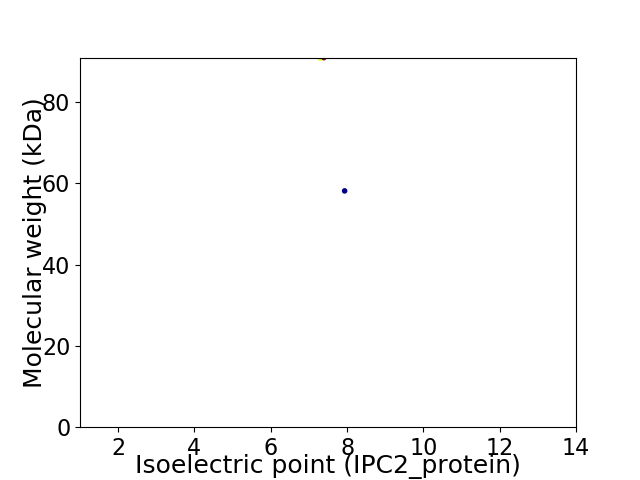
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
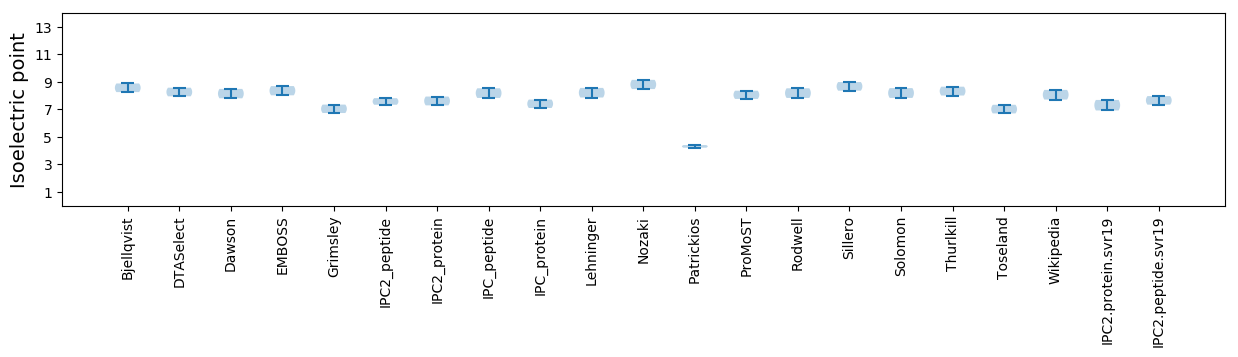
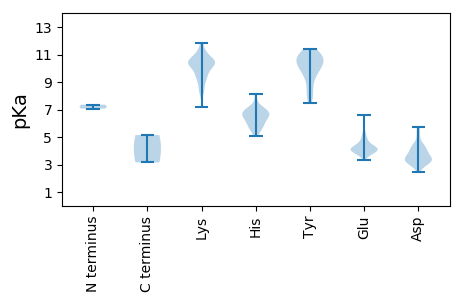
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W6EUV6|A0A1W6EUV6_9VIRU Protease OS=Salmovirus WFRC1 OX=1981916 PE=4 SV=1
MM1 pKa = 7.07FAWSRR6 pKa = 11.84PNSGLASAEE15 pKa = 3.56ILKK18 pKa = 9.88FRR20 pKa = 11.84KK21 pKa = 9.45PGFILSCEE29 pKa = 3.75QTARR33 pKa = 11.84LEE35 pKa = 4.12KK36 pKa = 10.64AKK38 pKa = 10.75EE39 pKa = 3.84KK40 pKa = 10.62VAEE43 pKa = 4.2ILCGATGCCRR53 pKa = 11.84RR54 pKa = 11.84KK55 pKa = 10.23RR56 pKa = 11.84LMDD59 pKa = 3.67LQVDD63 pKa = 4.41LSRR66 pKa = 11.84SPGYY70 pKa = 9.11PYY72 pKa = 10.66KK73 pKa = 10.58YY74 pKa = 10.11VYY76 pKa = 10.06RR77 pKa = 11.84SKK79 pKa = 10.94KK80 pKa = 10.26DD81 pKa = 3.37SLEE84 pKa = 4.09DD85 pKa = 4.16PIIYY89 pKa = 10.16DD90 pKa = 3.21RR91 pKa = 11.84VMTGNCWRR99 pKa = 11.84DD100 pKa = 3.32PAPLWDD106 pKa = 3.69AFGKK110 pKa = 10.58FEE112 pKa = 4.42ILPVEE117 pKa = 4.34KK118 pKa = 10.44ALKK121 pKa = 9.14RR122 pKa = 11.84CRR124 pKa = 11.84LITSPPVDD132 pKa = 3.2VALCVSMWYY141 pKa = 9.92EE142 pKa = 4.01DD143 pKa = 3.48QNEE146 pKa = 4.18RR147 pKa = 11.84LMSLCEE153 pKa = 3.84SSPYY157 pKa = 10.66GVGISKK163 pKa = 8.22FTGSWNRR170 pKa = 11.84LMSRR174 pKa = 11.84MPNVLWQCDD183 pKa = 3.56VSQFDD188 pKa = 3.34ACIRR192 pKa = 11.84PLLMSAVYY200 pKa = 9.93DD201 pKa = 3.6VRR203 pKa = 11.84EE204 pKa = 4.39LLFEE208 pKa = 5.3DD209 pKa = 4.16IPRR212 pKa = 11.84EE213 pKa = 3.96YY214 pKa = 11.4YY215 pKa = 10.16EE216 pKa = 6.62FRR218 pKa = 11.84DD219 pKa = 3.27QWLDD223 pKa = 3.31FLIDD227 pKa = 3.76GSFVDD232 pKa = 3.84HH233 pKa = 6.64HH234 pKa = 6.71TGDD237 pKa = 3.91LYY239 pKa = 11.38SVHH242 pKa = 6.85GGNKK246 pKa = 9.52SGSPNTSSDD255 pKa = 3.1NTIANMIVLCYY266 pKa = 11.14SMMSVGVNPMEE277 pKa = 4.44TPFVCYY283 pKa = 10.42GDD285 pKa = 5.51DD286 pKa = 4.15LLLCGSVPEE295 pKa = 4.92DD296 pKa = 2.78VWAAYY301 pKa = 9.22TSMGMIIKK309 pKa = 10.09PGACKK314 pKa = 9.72RR315 pKa = 11.84VMKK318 pKa = 10.41TEE320 pKa = 3.83VDD322 pKa = 4.15FLSCKK327 pKa = 8.34SLKK330 pKa = 10.36RR331 pKa = 11.84YY332 pKa = 7.81GQYY335 pKa = 10.22VPVWNSYY342 pKa = 8.47KK343 pKa = 10.31AAYY346 pKa = 10.77SMMVTDD352 pKa = 5.55SKK354 pKa = 11.83GEE356 pKa = 3.9LLRR359 pKa = 11.84GSRR362 pKa = 11.84FKK364 pKa = 11.19SLLLEE369 pKa = 4.16SLFVAEE375 pKa = 5.4FGVLSEE381 pKa = 4.24FARR384 pKa = 11.84LNGFFYY390 pKa = 10.86SRR392 pKa = 11.84SLLEE396 pKa = 4.62CIFFGMEE403 pKa = 3.83VEE405 pKa = 5.13DD406 pKa = 4.05KK407 pKa = 11.42KK408 pKa = 11.46EE409 pKa = 3.88MPGKK413 pKa = 10.15KK414 pKa = 9.74RR415 pKa = 11.84NTKK418 pKa = 8.57TKK420 pKa = 10.52QSLKK424 pKa = 10.35VPASRR429 pKa = 11.84PQKK432 pKa = 9.09QKK434 pKa = 9.23RR435 pKa = 11.84PRR437 pKa = 11.84RR438 pKa = 11.84RR439 pKa = 11.84RR440 pKa = 11.84QAGSRR445 pKa = 11.84TGQTDD450 pKa = 3.18DD451 pKa = 4.08SQWFMIHH458 pKa = 6.67RR459 pKa = 11.84EE460 pKa = 4.25AISTSTQSGVLFSSILHH477 pKa = 6.45PSLFPATPYY486 pKa = 9.02YY487 pKa = 9.72TRR489 pKa = 11.84CSNFTHH495 pKa = 7.13RR496 pKa = 11.84IEE498 pKa = 4.47RR499 pKa = 11.84SWEE502 pKa = 3.48FRR504 pKa = 11.84IMITTASFTGSRR516 pKa = 11.84VAVIPVNDD524 pKa = 4.5PSPDD528 pKa = 3.14AVMSEE533 pKa = 4.23TVAFQQVANGRR544 pKa = 11.84AAIATSTGASDD555 pKa = 3.19QRR557 pKa = 11.84SFVKK561 pKa = 10.05MYY563 pKa = 10.82GATTTLSNANPSVQSSLVGFSVGTFFVYY591 pKa = 10.91LLDD594 pKa = 4.17PPIGLAQGAALLVTVLARR612 pKa = 11.84VALTVVGPFTGFLTFEE628 pKa = 4.93ANPTDD633 pKa = 3.69HH634 pKa = 6.98GAPPVEE640 pKa = 4.48LKK642 pKa = 10.69ISDD645 pKa = 3.72VTKK648 pKa = 10.07DD649 pKa = 3.0RR650 pKa = 11.84SFITHH655 pKa = 6.39SKK657 pKa = 10.46SSYY660 pKa = 10.51LDD662 pKa = 3.21FGRR665 pKa = 11.84YY666 pKa = 8.2LAIEE670 pKa = 4.83GPSVTLDD677 pKa = 3.23SAEE680 pKa = 4.03WAVKK684 pKa = 10.34PEE686 pKa = 3.98FSRR689 pKa = 11.84IYY691 pKa = 10.96ASLSPPADD699 pKa = 2.57SWYY702 pKa = 10.81NNQDD706 pKa = 3.3EE707 pKa = 4.92KK708 pKa = 11.29IMPEE712 pKa = 4.77FYY714 pKa = 10.58TEE716 pKa = 3.93GDD718 pKa = 3.19LGGGVHH724 pKa = 7.0VMIGWPTIQDD734 pKa = 2.85ARR736 pKa = 11.84TFTVSPSSVVNGRR749 pKa = 11.84ALGFGSTDD757 pKa = 3.02SKK759 pKa = 11.64AKK761 pKa = 10.45KK762 pKa = 9.31KK763 pKa = 9.64WSEE766 pKa = 3.81TWSGGSEE773 pKa = 3.77SGQRR777 pKa = 11.84SIFFVAIGAQTVDD790 pKa = 4.42AMCCDD795 pKa = 3.33SFLGQRR801 pKa = 11.84LGALSLNSDD810 pKa = 3.76PSLLEE815 pKa = 3.81PP816 pKa = 5.14
MM1 pKa = 7.07FAWSRR6 pKa = 11.84PNSGLASAEE15 pKa = 3.56ILKK18 pKa = 9.88FRR20 pKa = 11.84KK21 pKa = 9.45PGFILSCEE29 pKa = 3.75QTARR33 pKa = 11.84LEE35 pKa = 4.12KK36 pKa = 10.64AKK38 pKa = 10.75EE39 pKa = 3.84KK40 pKa = 10.62VAEE43 pKa = 4.2ILCGATGCCRR53 pKa = 11.84RR54 pKa = 11.84KK55 pKa = 10.23RR56 pKa = 11.84LMDD59 pKa = 3.67LQVDD63 pKa = 4.41LSRR66 pKa = 11.84SPGYY70 pKa = 9.11PYY72 pKa = 10.66KK73 pKa = 10.58YY74 pKa = 10.11VYY76 pKa = 10.06RR77 pKa = 11.84SKK79 pKa = 10.94KK80 pKa = 10.26DD81 pKa = 3.37SLEE84 pKa = 4.09DD85 pKa = 4.16PIIYY89 pKa = 10.16DD90 pKa = 3.21RR91 pKa = 11.84VMTGNCWRR99 pKa = 11.84DD100 pKa = 3.32PAPLWDD106 pKa = 3.69AFGKK110 pKa = 10.58FEE112 pKa = 4.42ILPVEE117 pKa = 4.34KK118 pKa = 10.44ALKK121 pKa = 9.14RR122 pKa = 11.84CRR124 pKa = 11.84LITSPPVDD132 pKa = 3.2VALCVSMWYY141 pKa = 9.92EE142 pKa = 4.01DD143 pKa = 3.48QNEE146 pKa = 4.18RR147 pKa = 11.84LMSLCEE153 pKa = 3.84SSPYY157 pKa = 10.66GVGISKK163 pKa = 8.22FTGSWNRR170 pKa = 11.84LMSRR174 pKa = 11.84MPNVLWQCDD183 pKa = 3.56VSQFDD188 pKa = 3.34ACIRR192 pKa = 11.84PLLMSAVYY200 pKa = 9.93DD201 pKa = 3.6VRR203 pKa = 11.84EE204 pKa = 4.39LLFEE208 pKa = 5.3DD209 pKa = 4.16IPRR212 pKa = 11.84EE213 pKa = 3.96YY214 pKa = 11.4YY215 pKa = 10.16EE216 pKa = 6.62FRR218 pKa = 11.84DD219 pKa = 3.27QWLDD223 pKa = 3.31FLIDD227 pKa = 3.76GSFVDD232 pKa = 3.84HH233 pKa = 6.64HH234 pKa = 6.71TGDD237 pKa = 3.91LYY239 pKa = 11.38SVHH242 pKa = 6.85GGNKK246 pKa = 9.52SGSPNTSSDD255 pKa = 3.1NTIANMIVLCYY266 pKa = 11.14SMMSVGVNPMEE277 pKa = 4.44TPFVCYY283 pKa = 10.42GDD285 pKa = 5.51DD286 pKa = 4.15LLLCGSVPEE295 pKa = 4.92DD296 pKa = 2.78VWAAYY301 pKa = 9.22TSMGMIIKK309 pKa = 10.09PGACKK314 pKa = 9.72RR315 pKa = 11.84VMKK318 pKa = 10.41TEE320 pKa = 3.83VDD322 pKa = 4.15FLSCKK327 pKa = 8.34SLKK330 pKa = 10.36RR331 pKa = 11.84YY332 pKa = 7.81GQYY335 pKa = 10.22VPVWNSYY342 pKa = 8.47KK343 pKa = 10.31AAYY346 pKa = 10.77SMMVTDD352 pKa = 5.55SKK354 pKa = 11.83GEE356 pKa = 3.9LLRR359 pKa = 11.84GSRR362 pKa = 11.84FKK364 pKa = 11.19SLLLEE369 pKa = 4.16SLFVAEE375 pKa = 5.4FGVLSEE381 pKa = 4.24FARR384 pKa = 11.84LNGFFYY390 pKa = 10.86SRR392 pKa = 11.84SLLEE396 pKa = 4.62CIFFGMEE403 pKa = 3.83VEE405 pKa = 5.13DD406 pKa = 4.05KK407 pKa = 11.42KK408 pKa = 11.46EE409 pKa = 3.88MPGKK413 pKa = 10.15KK414 pKa = 9.74RR415 pKa = 11.84NTKK418 pKa = 8.57TKK420 pKa = 10.52QSLKK424 pKa = 10.35VPASRR429 pKa = 11.84PQKK432 pKa = 9.09QKK434 pKa = 9.23RR435 pKa = 11.84PRR437 pKa = 11.84RR438 pKa = 11.84RR439 pKa = 11.84RR440 pKa = 11.84QAGSRR445 pKa = 11.84TGQTDD450 pKa = 3.18DD451 pKa = 4.08SQWFMIHH458 pKa = 6.67RR459 pKa = 11.84EE460 pKa = 4.25AISTSTQSGVLFSSILHH477 pKa = 6.45PSLFPATPYY486 pKa = 9.02YY487 pKa = 9.72TRR489 pKa = 11.84CSNFTHH495 pKa = 7.13RR496 pKa = 11.84IEE498 pKa = 4.47RR499 pKa = 11.84SWEE502 pKa = 3.48FRR504 pKa = 11.84IMITTASFTGSRR516 pKa = 11.84VAVIPVNDD524 pKa = 4.5PSPDD528 pKa = 3.14AVMSEE533 pKa = 4.23TVAFQQVANGRR544 pKa = 11.84AAIATSTGASDD555 pKa = 3.19QRR557 pKa = 11.84SFVKK561 pKa = 10.05MYY563 pKa = 10.82GATTTLSNANPSVQSSLVGFSVGTFFVYY591 pKa = 10.91LLDD594 pKa = 4.17PPIGLAQGAALLVTVLARR612 pKa = 11.84VALTVVGPFTGFLTFEE628 pKa = 4.93ANPTDD633 pKa = 3.69HH634 pKa = 6.98GAPPVEE640 pKa = 4.48LKK642 pKa = 10.69ISDD645 pKa = 3.72VTKK648 pKa = 10.07DD649 pKa = 3.0RR650 pKa = 11.84SFITHH655 pKa = 6.39SKK657 pKa = 10.46SSYY660 pKa = 10.51LDD662 pKa = 3.21FGRR665 pKa = 11.84YY666 pKa = 8.2LAIEE670 pKa = 4.83GPSVTLDD677 pKa = 3.23SAEE680 pKa = 4.03WAVKK684 pKa = 10.34PEE686 pKa = 3.98FSRR689 pKa = 11.84IYY691 pKa = 10.96ASLSPPADD699 pKa = 2.57SWYY702 pKa = 10.81NNQDD706 pKa = 3.3EE707 pKa = 4.92KK708 pKa = 11.29IMPEE712 pKa = 4.77FYY714 pKa = 10.58TEE716 pKa = 3.93GDD718 pKa = 3.19LGGGVHH724 pKa = 7.0VMIGWPTIQDD734 pKa = 2.85ARR736 pKa = 11.84TFTVSPSSVVNGRR749 pKa = 11.84ALGFGSTDD757 pKa = 3.02SKK759 pKa = 11.64AKK761 pKa = 10.45KK762 pKa = 9.31KK763 pKa = 9.64WSEE766 pKa = 3.81TWSGGSEE773 pKa = 3.77SGQRR777 pKa = 11.84SIFFVAIGAQTVDD790 pKa = 4.42AMCCDD795 pKa = 3.33SFLGQRR801 pKa = 11.84LGALSLNSDD810 pKa = 3.76PSLLEE815 pKa = 3.81PP816 pKa = 5.14
Molecular weight: 90.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6EUV6|A0A1W6EUV6_9VIRU Protease OS=Salmovirus WFRC1 OX=1981916 PE=4 SV=1
MM1 pKa = 7.35SPKK4 pKa = 9.53RR5 pKa = 11.84DD6 pKa = 3.56QPVSVMTIAPYY17 pKa = 10.73VLLGVVGGFAYY28 pKa = 10.41AAMSSACSDD37 pKa = 4.54CEE39 pKa = 4.19HH40 pKa = 6.75TCQGTFTDD48 pKa = 5.74GMCLMCQKK56 pKa = 11.13GEE58 pKa = 4.11LQCEE62 pKa = 4.39SCEE65 pKa = 4.04QSAAEE70 pKa = 4.42TPTKK74 pKa = 8.51TRR76 pKa = 11.84KK77 pKa = 8.18RR78 pKa = 11.84RR79 pKa = 11.84KK80 pKa = 9.02YY81 pKa = 10.01KK82 pKa = 10.37KK83 pKa = 9.99EE84 pKa = 4.23CIASLWGEE92 pKa = 4.06VRR94 pKa = 11.84EE95 pKa = 5.25GFALWSPLAAVFGLHH110 pKa = 7.08HH111 pKa = 6.56YY112 pKa = 9.71RR113 pKa = 11.84DD114 pKa = 3.59AMLASKK120 pKa = 10.44ALEE123 pKa = 4.15GLEE126 pKa = 4.09GLVKK130 pKa = 10.4KK131 pKa = 10.29VWHH134 pKa = 6.75LGDD137 pKa = 3.37KK138 pKa = 9.62PKK140 pKa = 10.86PEE142 pKa = 4.52FSISDD147 pKa = 4.09CINRR151 pKa = 11.84VNKK154 pKa = 10.36RR155 pKa = 11.84ILSMGDD161 pKa = 2.96GYY163 pKa = 11.27EE164 pKa = 4.17ADD166 pKa = 4.44DD167 pKa = 4.98EE168 pKa = 4.71IMGYY172 pKa = 7.49STEE175 pKa = 4.23GSASRR180 pKa = 11.84ARR182 pKa = 11.84EE183 pKa = 3.56RR184 pKa = 11.84YY185 pKa = 7.92EE186 pKa = 3.77NSKK189 pKa = 10.27NRR191 pKa = 11.84YY192 pKa = 8.37DD193 pKa = 3.94RR194 pKa = 11.84EE195 pKa = 4.32GEE197 pKa = 3.91RR198 pKa = 11.84KK199 pKa = 9.32FIEE202 pKa = 4.1RR203 pKa = 11.84AKK205 pKa = 10.77LGFFGGCIDD214 pKa = 4.32KK215 pKa = 11.02VSNVCTNGLGFGLAAALFGCFIYY238 pKa = 10.11RR239 pKa = 11.84QRR241 pKa = 11.84VRR243 pKa = 11.84RR244 pKa = 11.84SLIYY248 pKa = 9.51GTDD251 pKa = 3.6AVKK254 pKa = 9.14KK255 pKa = 9.04TCDD258 pKa = 2.49KK259 pKa = 10.6WMKK262 pKa = 10.44KK263 pKa = 8.39DD264 pKa = 3.34TEE266 pKa = 4.22SRR268 pKa = 11.84ISGSHH273 pKa = 5.78TFTGCDD279 pKa = 3.41KK280 pKa = 10.43ICRR283 pKa = 11.84AVAQISTEE291 pKa = 4.07SGSVGTAFKK300 pKa = 10.57CGKK303 pKa = 8.51YY304 pKa = 9.44WITAGHH310 pKa = 5.08VVKK313 pKa = 10.93SGTAYY318 pKa = 11.01LHH320 pKa = 5.81TPSGKK325 pKa = 9.84EE326 pKa = 3.36ATEE329 pKa = 4.13VIKK332 pKa = 9.68FFPTANDD339 pKa = 5.28GITVMASKK347 pKa = 10.2YY348 pKa = 10.56SPMGLRR354 pKa = 11.84SVPVSVLTSALPGFVVGCQNGDD376 pKa = 3.26PHH378 pKa = 8.12LSTGLVEE385 pKa = 4.62PDD387 pKa = 2.97GTHH390 pKa = 5.52YY391 pKa = 11.04CSTTFGHH398 pKa = 6.2SGAPVVAHH406 pKa = 6.23NGVVVGVHH414 pKa = 5.64ILGDD418 pKa = 3.71EE419 pKa = 4.24DD420 pKa = 4.41VNGYY424 pKa = 10.75LPMTRR429 pKa = 11.84DD430 pKa = 3.07IISFLRR436 pKa = 11.84EE437 pKa = 4.0GPSAPVKK444 pKa = 10.34KK445 pKa = 10.22KK446 pKa = 10.63AVVPQDD452 pKa = 3.86VVSMAPITDD461 pKa = 4.31LLSQGMQQASVSNTLEE477 pKa = 4.01KK478 pKa = 10.46PGPCSHH484 pKa = 6.19GQGLIVDD491 pKa = 5.0LPLQRR496 pKa = 11.84SSSLEE501 pKa = 3.93SLDD504 pKa = 3.37SSSRR508 pKa = 11.84VSKK511 pKa = 10.53QHH513 pKa = 5.82DD514 pKa = 3.47LKK516 pKa = 10.91RR517 pKa = 11.84RR518 pKa = 11.84KK519 pKa = 9.95KK520 pKa = 9.28KK521 pKa = 7.19WQKK524 pKa = 10.75SSAVQPGVAAASVV537 pKa = 3.15
MM1 pKa = 7.35SPKK4 pKa = 9.53RR5 pKa = 11.84DD6 pKa = 3.56QPVSVMTIAPYY17 pKa = 10.73VLLGVVGGFAYY28 pKa = 10.41AAMSSACSDD37 pKa = 4.54CEE39 pKa = 4.19HH40 pKa = 6.75TCQGTFTDD48 pKa = 5.74GMCLMCQKK56 pKa = 11.13GEE58 pKa = 4.11LQCEE62 pKa = 4.39SCEE65 pKa = 4.04QSAAEE70 pKa = 4.42TPTKK74 pKa = 8.51TRR76 pKa = 11.84KK77 pKa = 8.18RR78 pKa = 11.84RR79 pKa = 11.84KK80 pKa = 9.02YY81 pKa = 10.01KK82 pKa = 10.37KK83 pKa = 9.99EE84 pKa = 4.23CIASLWGEE92 pKa = 4.06VRR94 pKa = 11.84EE95 pKa = 5.25GFALWSPLAAVFGLHH110 pKa = 7.08HH111 pKa = 6.56YY112 pKa = 9.71RR113 pKa = 11.84DD114 pKa = 3.59AMLASKK120 pKa = 10.44ALEE123 pKa = 4.15GLEE126 pKa = 4.09GLVKK130 pKa = 10.4KK131 pKa = 10.29VWHH134 pKa = 6.75LGDD137 pKa = 3.37KK138 pKa = 9.62PKK140 pKa = 10.86PEE142 pKa = 4.52FSISDD147 pKa = 4.09CINRR151 pKa = 11.84VNKK154 pKa = 10.36RR155 pKa = 11.84ILSMGDD161 pKa = 2.96GYY163 pKa = 11.27EE164 pKa = 4.17ADD166 pKa = 4.44DD167 pKa = 4.98EE168 pKa = 4.71IMGYY172 pKa = 7.49STEE175 pKa = 4.23GSASRR180 pKa = 11.84ARR182 pKa = 11.84EE183 pKa = 3.56RR184 pKa = 11.84YY185 pKa = 7.92EE186 pKa = 3.77NSKK189 pKa = 10.27NRR191 pKa = 11.84YY192 pKa = 8.37DD193 pKa = 3.94RR194 pKa = 11.84EE195 pKa = 4.32GEE197 pKa = 3.91RR198 pKa = 11.84KK199 pKa = 9.32FIEE202 pKa = 4.1RR203 pKa = 11.84AKK205 pKa = 10.77LGFFGGCIDD214 pKa = 4.32KK215 pKa = 11.02VSNVCTNGLGFGLAAALFGCFIYY238 pKa = 10.11RR239 pKa = 11.84QRR241 pKa = 11.84VRR243 pKa = 11.84RR244 pKa = 11.84SLIYY248 pKa = 9.51GTDD251 pKa = 3.6AVKK254 pKa = 9.14KK255 pKa = 9.04TCDD258 pKa = 2.49KK259 pKa = 10.6WMKK262 pKa = 10.44KK263 pKa = 8.39DD264 pKa = 3.34TEE266 pKa = 4.22SRR268 pKa = 11.84ISGSHH273 pKa = 5.78TFTGCDD279 pKa = 3.41KK280 pKa = 10.43ICRR283 pKa = 11.84AVAQISTEE291 pKa = 4.07SGSVGTAFKK300 pKa = 10.57CGKK303 pKa = 8.51YY304 pKa = 9.44WITAGHH310 pKa = 5.08VVKK313 pKa = 10.93SGTAYY318 pKa = 11.01LHH320 pKa = 5.81TPSGKK325 pKa = 9.84EE326 pKa = 3.36ATEE329 pKa = 4.13VIKK332 pKa = 9.68FFPTANDD339 pKa = 5.28GITVMASKK347 pKa = 10.2YY348 pKa = 10.56SPMGLRR354 pKa = 11.84SVPVSVLTSALPGFVVGCQNGDD376 pKa = 3.26PHH378 pKa = 8.12LSTGLVEE385 pKa = 4.62PDD387 pKa = 2.97GTHH390 pKa = 5.52YY391 pKa = 11.04CSTTFGHH398 pKa = 6.2SGAPVVAHH406 pKa = 6.23NGVVVGVHH414 pKa = 5.64ILGDD418 pKa = 3.71EE419 pKa = 4.24DD420 pKa = 4.41VNGYY424 pKa = 10.75LPMTRR429 pKa = 11.84DD430 pKa = 3.07IISFLRR436 pKa = 11.84EE437 pKa = 4.0GPSAPVKK444 pKa = 10.34KK445 pKa = 10.22KK446 pKa = 10.63AVVPQDD452 pKa = 3.86VVSMAPITDD461 pKa = 4.31LLSQGMQQASVSNTLEE477 pKa = 4.01KK478 pKa = 10.46PGPCSHH484 pKa = 6.19GQGLIVDD491 pKa = 5.0LPLQRR496 pKa = 11.84SSSLEE501 pKa = 3.93SLDD504 pKa = 3.37SSSRR508 pKa = 11.84VSKK511 pKa = 10.53QHH513 pKa = 5.82DD514 pKa = 3.47LKK516 pKa = 10.91RR517 pKa = 11.84RR518 pKa = 11.84KK519 pKa = 9.95KK520 pKa = 9.28KK521 pKa = 7.19WQKK524 pKa = 10.75SSAVQPGVAAASVV537 pKa = 3.15
Molecular weight: 58.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1353 |
537 |
816 |
676.5 |
74.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.095 ± 0.335 | 2.809 ± 0.454 |
5.174 ± 0.206 | 5.1 ± 0.071 |
4.582 ± 0.765 | 8.056 ± 1.012 |
1.7 ± 0.564 | 4.065 ± 0.096 |
5.913 ± 0.839 | 7.761 ± 0.541 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.956 ± 0.217 | 2.513 ± 0.289 |
5.174 ± 0.438 | 3.03 ± 0.084 |
5.691 ± 0.296 | 10.421 ± 0.343 |
5.617 ± 0.019 | 7.539 ± 0.407 |
1.626 ± 0.316 | 3.178 ± 0.239 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
