
Deinococcus puniceus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Deinococcus-Thermus; Deinococci; Deinococcales; Deinococcaceae; Deinococcus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
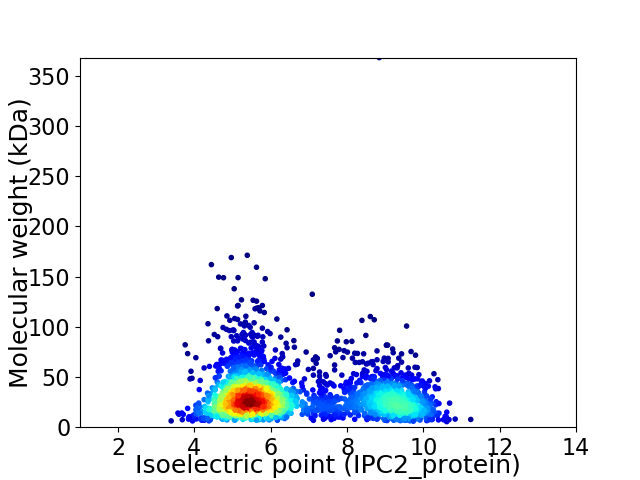
Virtual 2D-PAGE plot for 2606 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A172T6X9|A0A172T6X9_9DEIO Peptidyl-prolyl cis-trans isomerase OS=Deinococcus puniceus OX=1182568 GN=SU48_02320 PE=3 SV=1
MM1 pKa = 7.45LNVADD6 pKa = 4.73YY7 pKa = 11.41GDD9 pKa = 4.43APTAYY14 pKa = 10.16GNVAHH19 pKa = 6.85LPVLGWNGGTLGAGTNTIFPNTNTAPTFGLATQIPPTTALLGSVVDD65 pKa = 4.24VEE67 pKa = 4.54RR68 pKa = 11.84TSQANATATADD79 pKa = 3.79DD80 pKa = 4.8ANSIDD85 pKa = 4.98DD86 pKa = 4.03EE87 pKa = 5.99DD88 pKa = 4.91GVTMPASVMIGNVVTLPVAIGAAGLLSAWFDD119 pKa = 3.49WNRR122 pKa = 11.84DD123 pKa = 2.7GDD125 pKa = 3.88FADD128 pKa = 4.29AGEE131 pKa = 4.84QITSDD136 pKa = 3.11NSVSVGTTNLSVTVPTNAVAGTTFARR162 pKa = 11.84FRR164 pKa = 11.84ICTAAASCNSPTGVSPSGEE183 pKa = 4.07VEE185 pKa = 4.31DD186 pKa = 4.36YY187 pKa = 11.19SLVVMPTTADD197 pKa = 3.5LNINKK202 pKa = 8.23TGPSTVVQGSLATYY216 pKa = 8.48TITVWNKK223 pKa = 9.87GPGNSTGATISDD235 pKa = 4.03PVPSNLTNVNWTCAASDD252 pKa = 3.56TAACGIAGGSGNTINFVSGILPVNASATAPISGSYY287 pKa = 8.53LTLTVTGTATTSGTLINTASVTAAPGTTDD316 pKa = 3.73PVPGNNSSSQSTTVTAAMPVQPAINLKK343 pKa = 10.16KK344 pKa = 10.13YY345 pKa = 9.1VRR347 pKa = 11.84NVTTGTAFSEE357 pKa = 4.64TSTTARR363 pKa = 11.84PKK365 pKa = 10.66DD366 pKa = 3.31IIEE369 pKa = 4.06YY370 pKa = 10.18CITYY374 pKa = 10.35INAGGNAANFKK385 pKa = 9.62LTDD388 pKa = 3.66NVPAGMIVQLDD399 pKa = 4.27AYY401 pKa = 10.45AAGQGILWNSTSNTVSGNTTPAGTNLTNAPSDD433 pKa = 3.77DD434 pKa = 3.82AGTLTGADD442 pKa = 3.9TLPPIDD448 pKa = 3.88FTVHH452 pKa = 5.11GTNNTGLLTLDD463 pKa = 4.5LSATGLANLGKK474 pKa = 9.45GTVCFRR480 pKa = 11.84TQIPP484 pKa = 3.4
MM1 pKa = 7.45LNVADD6 pKa = 4.73YY7 pKa = 11.41GDD9 pKa = 4.43APTAYY14 pKa = 10.16GNVAHH19 pKa = 6.85LPVLGWNGGTLGAGTNTIFPNTNTAPTFGLATQIPPTTALLGSVVDD65 pKa = 4.24VEE67 pKa = 4.54RR68 pKa = 11.84TSQANATATADD79 pKa = 3.79DD80 pKa = 4.8ANSIDD85 pKa = 4.98DD86 pKa = 4.03EE87 pKa = 5.99DD88 pKa = 4.91GVTMPASVMIGNVVTLPVAIGAAGLLSAWFDD119 pKa = 3.49WNRR122 pKa = 11.84DD123 pKa = 2.7GDD125 pKa = 3.88FADD128 pKa = 4.29AGEE131 pKa = 4.84QITSDD136 pKa = 3.11NSVSVGTTNLSVTVPTNAVAGTTFARR162 pKa = 11.84FRR164 pKa = 11.84ICTAAASCNSPTGVSPSGEE183 pKa = 4.07VEE185 pKa = 4.31DD186 pKa = 4.36YY187 pKa = 11.19SLVVMPTTADD197 pKa = 3.5LNINKK202 pKa = 8.23TGPSTVVQGSLATYY216 pKa = 8.48TITVWNKK223 pKa = 9.87GPGNSTGATISDD235 pKa = 4.03PVPSNLTNVNWTCAASDD252 pKa = 3.56TAACGIAGGSGNTINFVSGILPVNASATAPISGSYY287 pKa = 8.53LTLTVTGTATTSGTLINTASVTAAPGTTDD316 pKa = 3.73PVPGNNSSSQSTTVTAAMPVQPAINLKK343 pKa = 10.16KK344 pKa = 10.13YY345 pKa = 9.1VRR347 pKa = 11.84NVTTGTAFSEE357 pKa = 4.64TSTTARR363 pKa = 11.84PKK365 pKa = 10.66DD366 pKa = 3.31IIEE369 pKa = 4.06YY370 pKa = 10.18CITYY374 pKa = 10.35INAGGNAANFKK385 pKa = 9.62LTDD388 pKa = 3.66NVPAGMIVQLDD399 pKa = 4.27AYY401 pKa = 10.45AAGQGILWNSTSNTVSGNTTPAGTNLTNAPSDD433 pKa = 3.77DD434 pKa = 3.82AGTLTGADD442 pKa = 3.9TLPPIDD448 pKa = 3.88FTVHH452 pKa = 5.11GTNNTGLLTLDD463 pKa = 4.5LSATGLANLGKK474 pKa = 9.45GTVCFRR480 pKa = 11.84TQIPP484 pKa = 3.4
Molecular weight: 48.72 kDa
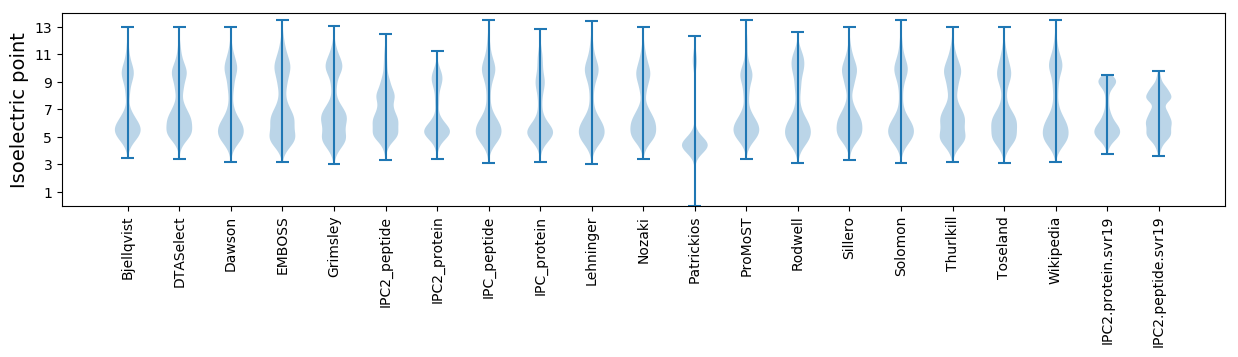
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A172TCG9|A0A172TCG9_9DEIO Transcriptional regulator OS=Deinococcus puniceus OX=1182568 GN=SU48_00050 PE=4 SV=1
MM1 pKa = 7.62LSLPAAKK8 pKa = 9.8PLTPARR14 pKa = 11.84RR15 pKa = 11.84PQPAIPRR22 pKa = 11.84QPLTRR27 pKa = 11.84RR28 pKa = 11.84TTAASQTTQWPVFRR42 pKa = 11.84PGHH45 pKa = 6.33KK46 pKa = 9.6LQQAHH51 pKa = 6.74RR52 pKa = 11.84PPHH55 pKa = 5.92RR56 pKa = 11.84KK57 pKa = 8.49PRR59 pKa = 11.84QARR62 pKa = 11.84GARR65 pKa = 11.84PFPP68 pKa = 4.4
MM1 pKa = 7.62LSLPAAKK8 pKa = 9.8PLTPARR14 pKa = 11.84RR15 pKa = 11.84PQPAIPRR22 pKa = 11.84QPLTRR27 pKa = 11.84RR28 pKa = 11.84TTAASQTTQWPVFRR42 pKa = 11.84PGHH45 pKa = 6.33KK46 pKa = 9.6LQQAHH51 pKa = 6.74RR52 pKa = 11.84PPHH55 pKa = 5.92RR56 pKa = 11.84KK57 pKa = 8.49PRR59 pKa = 11.84QARR62 pKa = 11.84GARR65 pKa = 11.84PFPP68 pKa = 4.4
Molecular weight: 7.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
801718 |
47 |
3650 |
307.6 |
33.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.726 ± 0.073 | 0.531 ± 0.013 |
5.126 ± 0.039 | 5.351 ± 0.058 |
3.169 ± 0.03 | 9.126 ± 0.057 |
2.052 ± 0.033 | 3.864 ± 0.043 |
2.757 ± 0.046 | 11.366 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.967 ± 0.025 | 2.704 ± 0.038 |
5.729 ± 0.042 | 3.684 ± 0.03 |
6.804 ± 0.048 | 5.11 ± 0.035 |
6.369 ± 0.064 | 7.912 ± 0.037 |
1.33 ± 0.022 | 2.325 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
