
Microviridae Bog5275_51
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.2
Get precalculated fractions of proteins
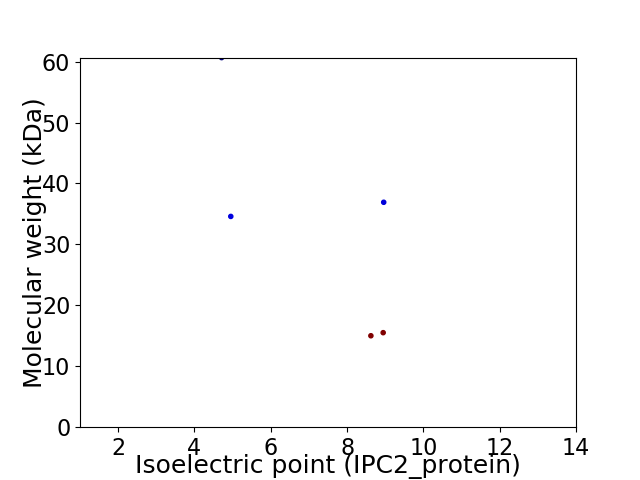
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
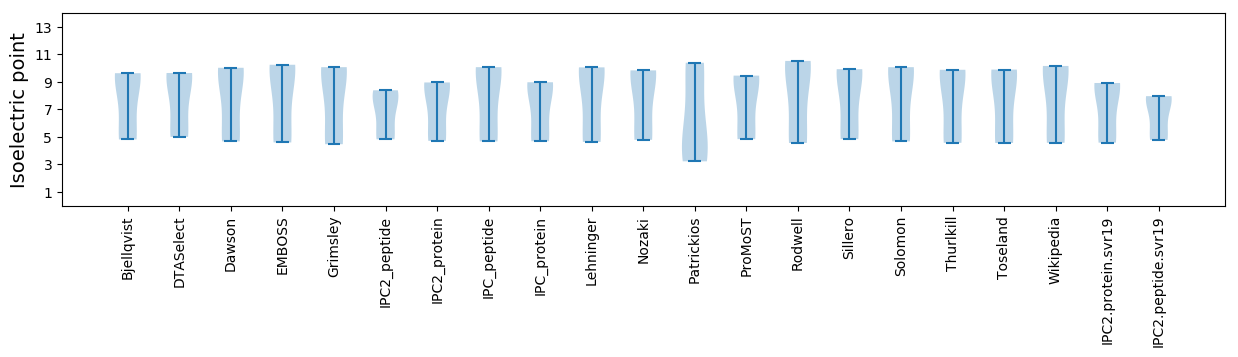
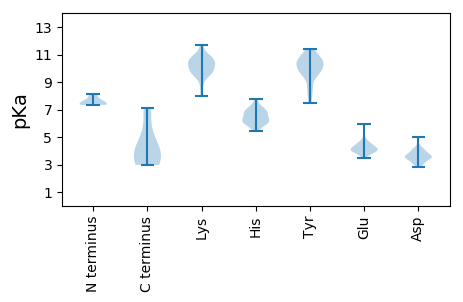
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G2UMC9|A0A0G2UMC9_9VIRU DNA pilot protein VP2 OS=Microviridae Bog5275_51 OX=1655648 PE=4 SV=1
MM1 pKa = 7.42SKK3 pKa = 10.21QRR5 pKa = 11.84IINTGQLNLTEE16 pKa = 5.06RR17 pKa = 11.84PTPNYY22 pKa = 8.79PHH24 pKa = 6.89NSFNLSHH31 pKa = 6.02SVKK34 pKa = 10.69LSFNYY39 pKa = 10.18GNLVPVMVMDD49 pKa = 3.81VVPGDD54 pKa = 3.63RR55 pKa = 11.84VHH57 pKa = 7.38IDD59 pKa = 3.23HH60 pKa = 7.16EE61 pKa = 4.57CLLRR65 pKa = 11.84YY66 pKa = 9.65QPMIAPPMQRR76 pKa = 11.84VKK78 pKa = 10.75VRR80 pKa = 11.84SEE82 pKa = 4.18SFFVPYY88 pKa = 10.3RR89 pKa = 11.84LLWPNFEE96 pKa = 4.4SAMGQRR102 pKa = 11.84DD103 pKa = 3.69VSDD106 pKa = 4.15PAPAFPTISVDD117 pKa = 3.49PEE119 pKa = 3.93VGISKK124 pKa = 10.57SSLANYY130 pKa = 10.12LGMPVLPVQPGGSTYY145 pKa = 10.76VYY147 pKa = 8.61PTMNAFRR154 pKa = 11.84FAAYY158 pKa = 10.11QKK160 pKa = 10.55IFADD164 pKa = 4.37WYY166 pKa = 10.67ADD168 pKa = 3.43EE169 pKa = 5.72DD170 pKa = 4.02LQIDD174 pKa = 4.22SVFVPLSDD182 pKa = 3.87GDD184 pKa = 3.73NGSIEE189 pKa = 4.02TSGYY193 pKa = 10.57GFLNQRR199 pKa = 11.84LYY201 pKa = 11.32DD202 pKa = 3.85LDD204 pKa = 4.14YY205 pKa = 9.97FTVAKK210 pKa = 9.58PWPQKK215 pKa = 10.05GSEE218 pKa = 4.24VVIPALNTQVPVSVIDD234 pKa = 3.38STAQQRR240 pKa = 11.84LYY242 pKa = 9.52QTVNGTYY249 pKa = 10.66ADD251 pKa = 3.55AGAPNIEE258 pKa = 4.65HH259 pKa = 7.77DD260 pKa = 4.33SMPIQAAFQDD270 pKa = 3.77GGGFLINLDD279 pKa = 3.89PKK281 pKa = 10.96GSLGINPADD290 pKa = 3.43WAGAAGTIEE299 pKa = 3.97QLRR302 pKa = 11.84VAEE305 pKa = 4.06AMQRR309 pKa = 11.84FLEE312 pKa = 4.47ADD314 pKa = 3.13SRR316 pKa = 11.84GGTRR320 pKa = 11.84YY321 pKa = 10.57GEE323 pKa = 4.43LIYY326 pKa = 11.08AHH328 pKa = 6.78FGVMNPDD335 pKa = 3.22ARR337 pKa = 11.84LQYY340 pKa = 10.43SEE342 pKa = 4.63YY343 pKa = 10.74LGSTSQQISFGEE355 pKa = 4.14VLNTTGTSTAPQGAMAGHH373 pKa = 6.76GVSAHH378 pKa = 5.9KK379 pKa = 9.69TNAPYY384 pKa = 10.12HH385 pKa = 6.3CNVTEE390 pKa = 4.24HH391 pKa = 6.46GVILTLMSVLPDD403 pKa = 2.98TGYY406 pKa = 10.12YY407 pKa = 9.68QGVPKK412 pKa = 10.53EE413 pKa = 3.91FFKK416 pKa = 11.1NDD418 pKa = 3.18RR419 pKa = 11.84LDD421 pKa = 3.98YY422 pKa = 10.6LWPEE426 pKa = 3.8YY427 pKa = 11.43AMLGEE432 pKa = 3.94QPINKK437 pKa = 9.7GEE439 pKa = 4.3VYY441 pKa = 10.19WDD443 pKa = 3.44VDD445 pKa = 3.55PSEE448 pKa = 5.92GSDD451 pKa = 3.53QNTQEE456 pKa = 3.7FGYY459 pKa = 10.35VGRR462 pKa = 11.84FNEE465 pKa = 4.56YY466 pKa = 7.51RR467 pKa = 11.84TQYY470 pKa = 11.05NRR472 pKa = 11.84VCGDD476 pKa = 3.33MAVSLDD482 pKa = 3.52FWQQDD487 pKa = 3.08RR488 pKa = 11.84KK489 pKa = 10.23FAGLPPLDD497 pKa = 3.86GADD500 pKa = 4.16FLTCVPDD507 pKa = 3.98DD508 pKa = 3.7ALNRR512 pKa = 11.84IFAVIDD518 pKa = 3.38ADD520 pKa = 4.04ADD522 pKa = 3.57HH523 pKa = 7.09LLAHH527 pKa = 7.06WWHH530 pKa = 6.09GVTVEE535 pKa = 3.99RR536 pKa = 11.84CLPRR540 pKa = 11.84LAMPSLL546 pKa = 3.86
MM1 pKa = 7.42SKK3 pKa = 10.21QRR5 pKa = 11.84IINTGQLNLTEE16 pKa = 5.06RR17 pKa = 11.84PTPNYY22 pKa = 8.79PHH24 pKa = 6.89NSFNLSHH31 pKa = 6.02SVKK34 pKa = 10.69LSFNYY39 pKa = 10.18GNLVPVMVMDD49 pKa = 3.81VVPGDD54 pKa = 3.63RR55 pKa = 11.84VHH57 pKa = 7.38IDD59 pKa = 3.23HH60 pKa = 7.16EE61 pKa = 4.57CLLRR65 pKa = 11.84YY66 pKa = 9.65QPMIAPPMQRR76 pKa = 11.84VKK78 pKa = 10.75VRR80 pKa = 11.84SEE82 pKa = 4.18SFFVPYY88 pKa = 10.3RR89 pKa = 11.84LLWPNFEE96 pKa = 4.4SAMGQRR102 pKa = 11.84DD103 pKa = 3.69VSDD106 pKa = 4.15PAPAFPTISVDD117 pKa = 3.49PEE119 pKa = 3.93VGISKK124 pKa = 10.57SSLANYY130 pKa = 10.12LGMPVLPVQPGGSTYY145 pKa = 10.76VYY147 pKa = 8.61PTMNAFRR154 pKa = 11.84FAAYY158 pKa = 10.11QKK160 pKa = 10.55IFADD164 pKa = 4.37WYY166 pKa = 10.67ADD168 pKa = 3.43EE169 pKa = 5.72DD170 pKa = 4.02LQIDD174 pKa = 4.22SVFVPLSDD182 pKa = 3.87GDD184 pKa = 3.73NGSIEE189 pKa = 4.02TSGYY193 pKa = 10.57GFLNQRR199 pKa = 11.84LYY201 pKa = 11.32DD202 pKa = 3.85LDD204 pKa = 4.14YY205 pKa = 9.97FTVAKK210 pKa = 9.58PWPQKK215 pKa = 10.05GSEE218 pKa = 4.24VVIPALNTQVPVSVIDD234 pKa = 3.38STAQQRR240 pKa = 11.84LYY242 pKa = 9.52QTVNGTYY249 pKa = 10.66ADD251 pKa = 3.55AGAPNIEE258 pKa = 4.65HH259 pKa = 7.77DD260 pKa = 4.33SMPIQAAFQDD270 pKa = 3.77GGGFLINLDD279 pKa = 3.89PKK281 pKa = 10.96GSLGINPADD290 pKa = 3.43WAGAAGTIEE299 pKa = 3.97QLRR302 pKa = 11.84VAEE305 pKa = 4.06AMQRR309 pKa = 11.84FLEE312 pKa = 4.47ADD314 pKa = 3.13SRR316 pKa = 11.84GGTRR320 pKa = 11.84YY321 pKa = 10.57GEE323 pKa = 4.43LIYY326 pKa = 11.08AHH328 pKa = 6.78FGVMNPDD335 pKa = 3.22ARR337 pKa = 11.84LQYY340 pKa = 10.43SEE342 pKa = 4.63YY343 pKa = 10.74LGSTSQQISFGEE355 pKa = 4.14VLNTTGTSTAPQGAMAGHH373 pKa = 6.76GVSAHH378 pKa = 5.9KK379 pKa = 9.69TNAPYY384 pKa = 10.12HH385 pKa = 6.3CNVTEE390 pKa = 4.24HH391 pKa = 6.46GVILTLMSVLPDD403 pKa = 2.98TGYY406 pKa = 10.12YY407 pKa = 9.68QGVPKK412 pKa = 10.53EE413 pKa = 3.91FFKK416 pKa = 11.1NDD418 pKa = 3.18RR419 pKa = 11.84LDD421 pKa = 3.98YY422 pKa = 10.6LWPEE426 pKa = 3.8YY427 pKa = 11.43AMLGEE432 pKa = 3.94QPINKK437 pKa = 9.7GEE439 pKa = 4.3VYY441 pKa = 10.19WDD443 pKa = 3.44VDD445 pKa = 3.55PSEE448 pKa = 5.92GSDD451 pKa = 3.53QNTQEE456 pKa = 3.7FGYY459 pKa = 10.35VGRR462 pKa = 11.84FNEE465 pKa = 4.56YY466 pKa = 7.51RR467 pKa = 11.84TQYY470 pKa = 11.05NRR472 pKa = 11.84VCGDD476 pKa = 3.33MAVSLDD482 pKa = 3.52FWQQDD487 pKa = 3.08RR488 pKa = 11.84KK489 pKa = 10.23FAGLPPLDD497 pKa = 3.86GADD500 pKa = 4.16FLTCVPDD507 pKa = 3.98DD508 pKa = 3.7ALNRR512 pKa = 11.84IFAVIDD518 pKa = 3.38ADD520 pKa = 4.04ADD522 pKa = 3.57HH523 pKa = 7.09LLAHH527 pKa = 7.06WWHH530 pKa = 6.09GVTVEE535 pKa = 3.99RR536 pKa = 11.84CLPRR540 pKa = 11.84LAMPSLL546 pKa = 3.86
Molecular weight: 60.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G2UK08|A0A0G2UK08_9VIRU Major capsid protein VP1 OS=Microviridae Bog5275_51 OX=1655648 PE=3 SV=1
MM1 pKa = 7.61AKK3 pKa = 10.16AIFAAPVPLIRR14 pKa = 11.84NHH16 pKa = 6.14FNYY19 pKa = 10.42DD20 pKa = 3.23YY21 pKa = 11.31AEE23 pKa = 4.13TFGRR27 pKa = 11.84VSPNKK32 pKa = 10.54SMTVPGAVKK41 pKa = 10.3SLAWMIDD48 pKa = 2.83RR49 pKa = 11.84FTRR52 pKa = 11.84NLAMPPAPAAFNTEE66 pKa = 4.18LDD68 pKa = 3.69LSPFDD73 pKa = 3.77RR74 pKa = 11.84LNVFEE79 pKa = 5.94KK80 pKa = 10.27IDD82 pKa = 3.48LSRR85 pKa = 11.84AYY87 pKa = 9.57KK88 pKa = 10.19RR89 pKa = 11.84RR90 pKa = 11.84ALSLRR95 pKa = 11.84KK96 pKa = 9.45QLDD99 pKa = 3.54DD100 pKa = 5.0KK101 pKa = 11.35RR102 pKa = 11.84QADD105 pKa = 3.52ANAEE109 pKa = 3.81QEE111 pKa = 4.14RR112 pKa = 11.84FIEE115 pKa = 3.95EE116 pKa = 3.9RR117 pKa = 11.84AALRR121 pKa = 11.84AIEE124 pKa = 4.02LSKK127 pKa = 10.6LAKK130 pKa = 9.64DD131 pKa = 4.09AKK133 pKa = 10.65DD134 pKa = 3.62GQAA137 pKa = 3.86
MM1 pKa = 7.61AKK3 pKa = 10.16AIFAAPVPLIRR14 pKa = 11.84NHH16 pKa = 6.14FNYY19 pKa = 10.42DD20 pKa = 3.23YY21 pKa = 11.31AEE23 pKa = 4.13TFGRR27 pKa = 11.84VSPNKK32 pKa = 10.54SMTVPGAVKK41 pKa = 10.3SLAWMIDD48 pKa = 2.83RR49 pKa = 11.84FTRR52 pKa = 11.84NLAMPPAPAAFNTEE66 pKa = 4.18LDD68 pKa = 3.69LSPFDD73 pKa = 3.77RR74 pKa = 11.84LNVFEE79 pKa = 5.94KK80 pKa = 10.27IDD82 pKa = 3.48LSRR85 pKa = 11.84AYY87 pKa = 9.57KK88 pKa = 10.19RR89 pKa = 11.84RR90 pKa = 11.84ALSLRR95 pKa = 11.84KK96 pKa = 9.45QLDD99 pKa = 3.54DD100 pKa = 5.0KK101 pKa = 11.35RR102 pKa = 11.84QADD105 pKa = 3.52ANAEE109 pKa = 3.81QEE111 pKa = 4.14RR112 pKa = 11.84FIEE115 pKa = 3.95EE116 pKa = 3.9RR117 pKa = 11.84AALRR121 pKa = 11.84AIEE124 pKa = 4.02LSKK127 pKa = 10.6LAKK130 pKa = 9.64DD131 pKa = 4.09AKK133 pKa = 10.65DD134 pKa = 3.62GQAA137 pKa = 3.86
Molecular weight: 15.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1480 |
137 |
546 |
296.0 |
32.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.757 ± 2.588 | 0.811 ± 0.366 |
6.149 ± 0.491 | 3.784 ± 0.541 |
4.122 ± 0.381 | 6.959 ± 0.754 |
2.095 ± 0.47 | 3.986 ± 0.635 |
3.986 ± 0.809 | 7.973 ± 0.42 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.905 ± 0.149 | 5.608 ± 1.102 |
6.081 ± 0.597 | 5.405 ± 0.654 |
5.473 ± 1.039 | 6.081 ± 0.383 |
5.405 ± 0.804 | 6.486 ± 0.725 |
1.351 ± 0.36 | 3.581 ± 0.853 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
