
Bat coronavirus HKU9-4
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Nobecovirus; Rousettus bat coronavirus HKU9
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
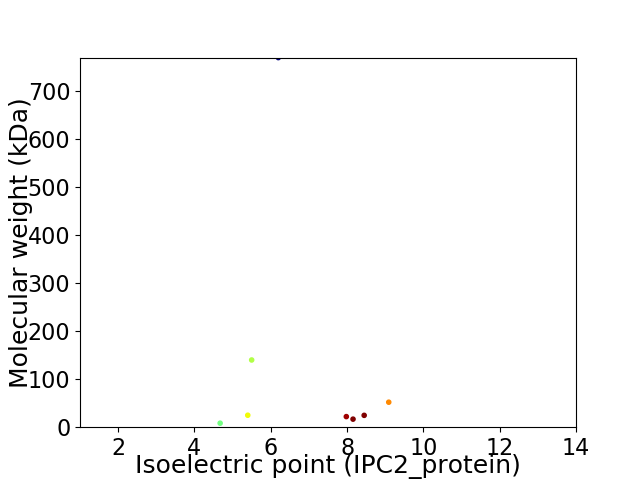
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A3EXJ3|A3EXJ3_BCHK9 Membrane protein OS=Bat coronavirus HKU9-4 OX=424370 GN=M PE=3 SV=1
MM1 pKa = 7.32YY2 pKa = 9.76EE3 pKa = 4.44LVSADD8 pKa = 3.47TSVVIANVLVIIILCLFVVIVGCALLLILQFILGTCGCLFSVICKK53 pKa = 8.12PTILVYY59 pKa = 11.09NKK61 pKa = 9.81FKK63 pKa = 11.29NEE65 pKa = 3.87SLLNEE70 pKa = 4.09QEE72 pKa = 4.15EE73 pKa = 4.57LLVV76 pKa = 3.8
MM1 pKa = 7.32YY2 pKa = 9.76EE3 pKa = 4.44LVSADD8 pKa = 3.47TSVVIANVLVIIILCLFVVIVGCALLLILQFILGTCGCLFSVICKK53 pKa = 8.12PTILVYY59 pKa = 11.09NKK61 pKa = 9.81FKK63 pKa = 11.29NEE65 pKa = 3.87SLLNEE70 pKa = 4.09QEE72 pKa = 4.15EE73 pKa = 4.57LLVV76 pKa = 3.8
Molecular weight: 8.38 kDa
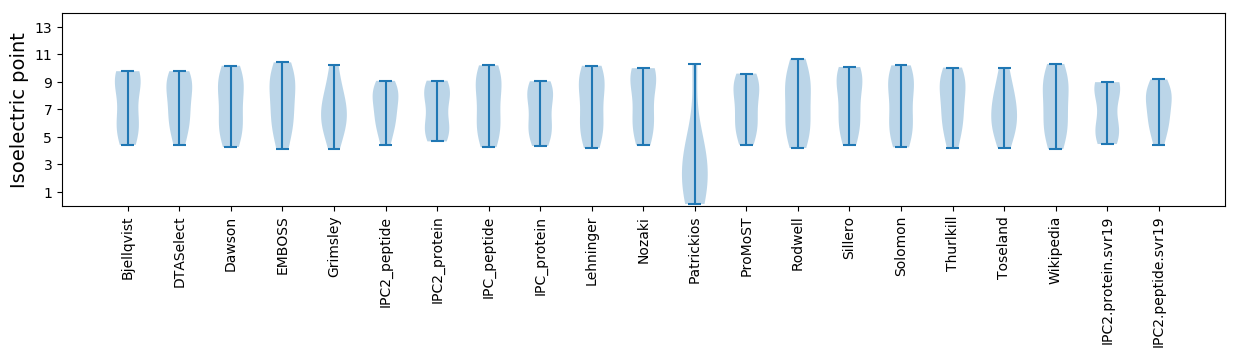
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A3EXJ5|A3EXJ5_BCHK9 Uncharacterized protein NS7a OS=Bat coronavirus HKU9-4 OX=424370 GN=NS7a PE=4 SV=1
MM1 pKa = 7.62SGRR4 pKa = 11.84NKK6 pKa = 9.51PRR8 pKa = 11.84PGSQSPKK15 pKa = 8.6VTFKK19 pKa = 11.01QEE21 pKa = 3.89SDD23 pKa = 3.52GSDD26 pKa = 3.37SEE28 pKa = 4.52SEE30 pKa = 3.89RR31 pKa = 11.84RR32 pKa = 11.84NGNRR36 pKa = 11.84SGARR40 pKa = 11.84PKK42 pKa = 10.88NNNNRR47 pKa = 11.84GGNSKK52 pKa = 10.38PEE54 pKa = 4.09KK55 pKa = 9.82PKK57 pKa = 10.53AAPPQNVSWFAPLVQTGKK75 pKa = 10.3QDD77 pKa = 3.28LRR79 pKa = 11.84FPRR82 pKa = 11.84GEE84 pKa = 4.24GVPISQGVDD93 pKa = 3.01PVYY96 pKa = 11.0LHH98 pKa = 6.8GYY100 pKa = 6.03WVRR103 pKa = 11.84TQRR106 pKa = 11.84NFQKK110 pKa = 10.75GGKK113 pKa = 6.6TVQANPRR120 pKa = 11.84WYY122 pKa = 10.03FYY124 pKa = 9.31YY125 pKa = 10.37TGTGRR130 pKa = 11.84HH131 pKa = 6.14ADD133 pKa = 3.75LRR135 pKa = 11.84WGQKK139 pKa = 10.17NPDD142 pKa = 4.47LIWVGEE148 pKa = 4.16DD149 pKa = 3.05GANINRR155 pKa = 11.84IGDD158 pKa = 3.47MGTRR162 pKa = 11.84NPNNDD167 pKa = 2.78SAIPVQFGSGIPKK180 pKa = 10.05GFYY183 pKa = 10.47AEE185 pKa = 4.29GRR187 pKa = 11.84NSRR190 pKa = 11.84GNSRR194 pKa = 11.84NSSRR198 pKa = 11.84NSSRR202 pKa = 11.84ASTRR206 pKa = 11.84GNSRR210 pKa = 11.84ANSRR214 pKa = 11.84GASPGRR220 pKa = 11.84NTPSGGSNSEE230 pKa = 3.14PWMAYY235 pKa = 9.67LVQKK239 pKa = 11.06LEE241 pKa = 4.02ALEE244 pKa = 4.26TKK246 pKa = 10.77VNGNKK251 pKa = 9.83SEE253 pKa = 4.25TKK255 pKa = 10.71APVQVTKK262 pKa = 10.97NAAAEE267 pKa = 3.96NAKK270 pKa = 10.26KK271 pKa = 10.1LRR273 pKa = 11.84HH274 pKa = 5.99KK275 pKa = 8.81RR276 pKa = 11.84TPHH279 pKa = 6.14KK280 pKa = 10.82GSGVTANFGRR290 pKa = 11.84RR291 pKa = 11.84GPGEE295 pKa = 3.93LEE297 pKa = 4.13GNFGDD302 pKa = 5.97LEE304 pKa = 4.15MLKK307 pKa = 10.67LGADD311 pKa = 3.89DD312 pKa = 4.27PRR314 pKa = 11.84FPAVAQMAPNVAAFMFMSHH333 pKa = 6.92FSTRR337 pKa = 11.84DD338 pKa = 3.19EE339 pKa = 5.27EE340 pKa = 4.27DD341 pKa = 3.94ALWLNYY347 pKa = 9.88RR348 pKa = 11.84GAIKK352 pKa = 10.43LPKK355 pKa = 9.56SDD357 pKa = 4.52PNFEE361 pKa = 3.93QWTKK365 pKa = 11.05LLEE368 pKa = 4.49EE369 pKa = 4.59NLNAYY374 pKa = 7.5KK375 pKa = 10.13TFPPPAPKK383 pKa = 9.39KK384 pKa = 9.89DD385 pKa = 3.35KK386 pKa = 10.55KK387 pKa = 10.65KK388 pKa = 10.77KK389 pKa = 9.84EE390 pKa = 3.91EE391 pKa = 4.04ASQEE395 pKa = 3.67IAIFEE400 pKa = 4.41DD401 pKa = 4.01ASTGTDD407 pKa = 3.36QQIVKK412 pKa = 9.86VWVKK416 pKa = 11.32DD417 pKa = 3.54EE418 pKa = 4.17GAQTDD423 pKa = 4.21EE424 pKa = 4.23EE425 pKa = 4.73WLGGDD430 pKa = 3.14DD431 pKa = 3.39TVYY434 pKa = 10.84EE435 pKa = 4.38EE436 pKa = 4.62EE437 pKa = 4.59DD438 pKa = 4.11DD439 pKa = 3.96RR440 pKa = 11.84PKK442 pKa = 9.38TQRR445 pKa = 11.84RR446 pKa = 11.84HH447 pKa = 5.15KK448 pKa = 10.06KK449 pKa = 9.76RR450 pKa = 11.84NSTASRR456 pKa = 11.84VTIADD461 pKa = 3.43PMNATSEE468 pKa = 4.26RR469 pKa = 11.84SS470 pKa = 3.23
MM1 pKa = 7.62SGRR4 pKa = 11.84NKK6 pKa = 9.51PRR8 pKa = 11.84PGSQSPKK15 pKa = 8.6VTFKK19 pKa = 11.01QEE21 pKa = 3.89SDD23 pKa = 3.52GSDD26 pKa = 3.37SEE28 pKa = 4.52SEE30 pKa = 3.89RR31 pKa = 11.84RR32 pKa = 11.84NGNRR36 pKa = 11.84SGARR40 pKa = 11.84PKK42 pKa = 10.88NNNNRR47 pKa = 11.84GGNSKK52 pKa = 10.38PEE54 pKa = 4.09KK55 pKa = 9.82PKK57 pKa = 10.53AAPPQNVSWFAPLVQTGKK75 pKa = 10.3QDD77 pKa = 3.28LRR79 pKa = 11.84FPRR82 pKa = 11.84GEE84 pKa = 4.24GVPISQGVDD93 pKa = 3.01PVYY96 pKa = 11.0LHH98 pKa = 6.8GYY100 pKa = 6.03WVRR103 pKa = 11.84TQRR106 pKa = 11.84NFQKK110 pKa = 10.75GGKK113 pKa = 6.6TVQANPRR120 pKa = 11.84WYY122 pKa = 10.03FYY124 pKa = 9.31YY125 pKa = 10.37TGTGRR130 pKa = 11.84HH131 pKa = 6.14ADD133 pKa = 3.75LRR135 pKa = 11.84WGQKK139 pKa = 10.17NPDD142 pKa = 4.47LIWVGEE148 pKa = 4.16DD149 pKa = 3.05GANINRR155 pKa = 11.84IGDD158 pKa = 3.47MGTRR162 pKa = 11.84NPNNDD167 pKa = 2.78SAIPVQFGSGIPKK180 pKa = 10.05GFYY183 pKa = 10.47AEE185 pKa = 4.29GRR187 pKa = 11.84NSRR190 pKa = 11.84GNSRR194 pKa = 11.84NSSRR198 pKa = 11.84NSSRR202 pKa = 11.84ASTRR206 pKa = 11.84GNSRR210 pKa = 11.84ANSRR214 pKa = 11.84GASPGRR220 pKa = 11.84NTPSGGSNSEE230 pKa = 3.14PWMAYY235 pKa = 9.67LVQKK239 pKa = 11.06LEE241 pKa = 4.02ALEE244 pKa = 4.26TKK246 pKa = 10.77VNGNKK251 pKa = 9.83SEE253 pKa = 4.25TKK255 pKa = 10.71APVQVTKK262 pKa = 10.97NAAAEE267 pKa = 3.96NAKK270 pKa = 10.26KK271 pKa = 10.1LRR273 pKa = 11.84HH274 pKa = 5.99KK275 pKa = 8.81RR276 pKa = 11.84TPHH279 pKa = 6.14KK280 pKa = 10.82GSGVTANFGRR290 pKa = 11.84RR291 pKa = 11.84GPGEE295 pKa = 3.93LEE297 pKa = 4.13GNFGDD302 pKa = 5.97LEE304 pKa = 4.15MLKK307 pKa = 10.67LGADD311 pKa = 3.89DD312 pKa = 4.27PRR314 pKa = 11.84FPAVAQMAPNVAAFMFMSHH333 pKa = 6.92FSTRR337 pKa = 11.84DD338 pKa = 3.19EE339 pKa = 5.27EE340 pKa = 4.27DD341 pKa = 3.94ALWLNYY347 pKa = 9.88RR348 pKa = 11.84GAIKK352 pKa = 10.43LPKK355 pKa = 9.56SDD357 pKa = 4.52PNFEE361 pKa = 3.93QWTKK365 pKa = 11.05LLEE368 pKa = 4.49EE369 pKa = 4.59NLNAYY374 pKa = 7.5KK375 pKa = 10.13TFPPPAPKK383 pKa = 9.39KK384 pKa = 9.89DD385 pKa = 3.35KK386 pKa = 10.55KK387 pKa = 10.65KK388 pKa = 10.77KK389 pKa = 9.84EE390 pKa = 3.91EE391 pKa = 4.04ASQEE395 pKa = 3.67IAIFEE400 pKa = 4.41DD401 pKa = 4.01ASTGTDD407 pKa = 3.36QQIVKK412 pKa = 9.86VWVKK416 pKa = 11.32DD417 pKa = 3.54EE418 pKa = 4.17GAQTDD423 pKa = 4.21EE424 pKa = 4.23EE425 pKa = 4.73WLGGDD430 pKa = 3.14DD431 pKa = 3.39TVYY434 pKa = 10.84EE435 pKa = 4.38EE436 pKa = 4.62EE437 pKa = 4.59DD438 pKa = 4.11DD439 pKa = 3.96RR440 pKa = 11.84PKK442 pKa = 9.38TQRR445 pKa = 11.84RR446 pKa = 11.84HH447 pKa = 5.15KK448 pKa = 10.06KK449 pKa = 9.76RR450 pKa = 11.84NSTASRR456 pKa = 11.84VTIADD461 pKa = 3.43PMNATSEE468 pKa = 4.26RR469 pKa = 11.84SS470 pKa = 3.23
Molecular weight: 52.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9540 |
76 |
6925 |
1192.5 |
132.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.505 ± 0.313 | 3.092 ± 0.342 |
5.273 ± 0.781 | 3.899 ± 0.343 |
4.486 ± 0.389 | 6.31 ± 0.402 |
1.824 ± 0.281 | 4.329 ± 0.655 |
4.801 ± 0.661 | 9.549 ± 0.724 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.4 ± 0.364 | 5.042 ± 0.848 |
4.675 ± 0.334 | 3.365 ± 0.388 |
4.308 ± 0.511 | 6.813 ± 0.52 |
6.52 ± 0.342 | 9.696 ± 0.797 |
1.31 ± 0.28 | 4.79 ± 0.553 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
