
Strawberry vein banding virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Caulimovirus
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
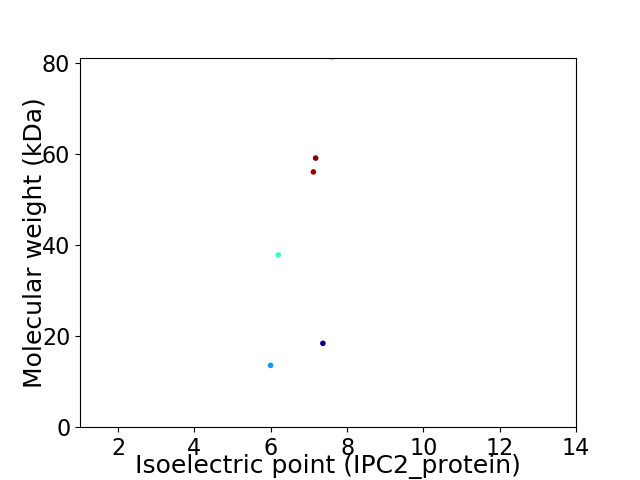
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q88441|Q88441_9VIRU Coat protein OS=Strawberry vein banding virus OX=47903 GN=ORF IV PE=3 SV=1
MM1 pKa = 7.67GSVTQKK7 pKa = 8.87TEE9 pKa = 4.06RR10 pKa = 11.84IYY12 pKa = 11.37DD13 pKa = 3.52EE14 pKa = 4.02MLKK17 pKa = 9.47MEE19 pKa = 4.36EE20 pKa = 3.9RR21 pKa = 11.84FSRR24 pKa = 11.84LEE26 pKa = 3.93QEE28 pKa = 4.46QQDD31 pKa = 4.0HH32 pKa = 6.3FSEE35 pKa = 5.62KK36 pKa = 9.3IDD38 pKa = 3.51QASAILLAEE47 pKa = 3.79IRR49 pKa = 11.84KK50 pKa = 8.8IQSKK54 pKa = 11.17LEE56 pKa = 3.94TCDD59 pKa = 3.53CNKK62 pKa = 10.55EE63 pKa = 3.83ILDD66 pKa = 3.79ALKK69 pKa = 10.89AQDD72 pKa = 3.21RR73 pKa = 11.84TKK75 pKa = 11.18NKK77 pKa = 10.31GSGPSNLDD85 pKa = 3.53ALDD88 pKa = 4.51LAHH91 pKa = 7.49KK92 pKa = 10.58DD93 pKa = 3.49NSPKK97 pKa = 10.33RR98 pKa = 11.84NPIRR102 pKa = 11.84GTEE105 pKa = 3.71NWHH108 pKa = 5.77PQDD111 pKa = 5.06LKK113 pKa = 9.48LTKK116 pKa = 8.98WW117 pKa = 3.24
MM1 pKa = 7.67GSVTQKK7 pKa = 8.87TEE9 pKa = 4.06RR10 pKa = 11.84IYY12 pKa = 11.37DD13 pKa = 3.52EE14 pKa = 4.02MLKK17 pKa = 9.47MEE19 pKa = 4.36EE20 pKa = 3.9RR21 pKa = 11.84FSRR24 pKa = 11.84LEE26 pKa = 3.93QEE28 pKa = 4.46QQDD31 pKa = 4.0HH32 pKa = 6.3FSEE35 pKa = 5.62KK36 pKa = 9.3IDD38 pKa = 3.51QASAILLAEE47 pKa = 3.79IRR49 pKa = 11.84KK50 pKa = 8.8IQSKK54 pKa = 11.17LEE56 pKa = 3.94TCDD59 pKa = 3.53CNKK62 pKa = 10.55EE63 pKa = 3.83ILDD66 pKa = 3.79ALKK69 pKa = 10.89AQDD72 pKa = 3.21RR73 pKa = 11.84TKK75 pKa = 11.18NKK77 pKa = 10.31GSGPSNLDD85 pKa = 3.53ALDD88 pKa = 4.51LAHH91 pKa = 7.49KK92 pKa = 10.58DD93 pKa = 3.49NSPKK97 pKa = 10.33RR98 pKa = 11.84NPIRR102 pKa = 11.84GTEE105 pKa = 3.71NWHH108 pKa = 5.77PQDD111 pKa = 5.06LKK113 pKa = 9.48LTKK116 pKa = 8.98WW117 pKa = 3.24
Molecular weight: 13.57 kDa
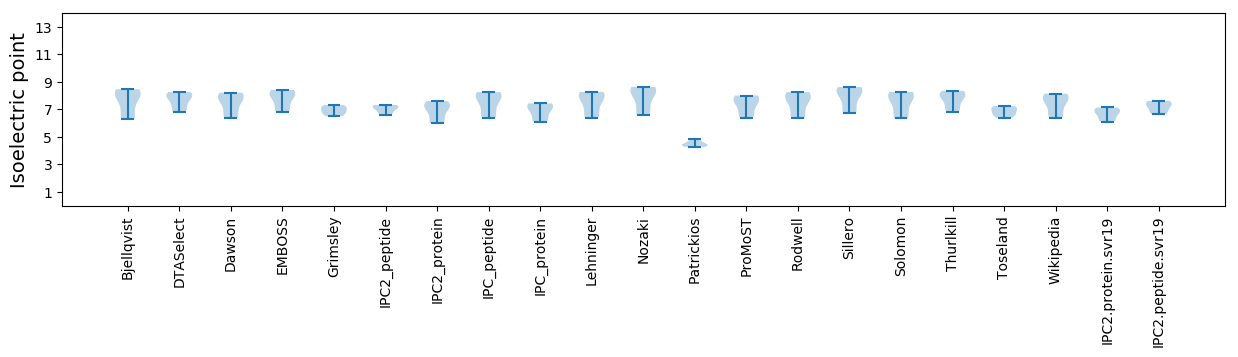
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q88443|Q88443_9VIRU ORF VI protein OS=Strawberry vein banding virus OX=47903 GN=ORF VI PE=4 SV=1
MM1 pKa = 7.66EE2 pKa = 5.88NNLQLLEE9 pKa = 4.22EE10 pKa = 4.68LNHH13 pKa = 7.69LEE15 pKa = 4.73EE16 pKa = 4.64GWEE19 pKa = 3.95MLTDD23 pKa = 3.8EE24 pKa = 5.47DD25 pKa = 6.11FKK27 pKa = 11.43LQQNEE32 pKa = 4.33DD33 pKa = 3.81EE34 pKa = 4.43EE35 pKa = 4.46QLLEE39 pKa = 4.29SVDD42 pKa = 3.62SEE44 pKa = 4.44EE45 pKa = 5.18RR46 pKa = 11.84FSTLTKK52 pKa = 10.01TNPNSIYY59 pKa = 10.23IRR61 pKa = 11.84GNFYY65 pKa = 10.26FKK67 pKa = 10.67GYY69 pKa = 10.34KK70 pKa = 9.7KK71 pKa = 10.74YY72 pKa = 11.06SLDD75 pKa = 3.79LYY77 pKa = 10.59VDD79 pKa = 4.32TGASMCTANKK89 pKa = 10.02HH90 pKa = 5.49VIPEE94 pKa = 4.23EE95 pKa = 3.76FWVNAKK101 pKa = 10.44NPIRR105 pKa = 11.84ARR107 pKa = 11.84IANDD111 pKa = 3.53SIMTFNKK118 pKa = 9.43VAEE121 pKa = 4.13LMQVQIADD129 pKa = 3.66EE130 pKa = 4.3TFIIPTLYY138 pKa = 10.45QATTKK143 pKa = 11.15GDD145 pKa = 3.12ITLGNNFCRR154 pKa = 11.84LYY156 pKa = 10.98EE157 pKa = 4.01PFVQYY162 pKa = 11.36KK163 pKa = 11.03DD164 pKa = 3.49MITFHH169 pKa = 6.96KK170 pKa = 10.33DD171 pKa = 2.48GRR173 pKa = 11.84AVSTKK178 pKa = 10.15KK179 pKa = 8.48VTKK182 pKa = 10.07AYY184 pKa = 10.35FHH186 pKa = 6.73GLPGFLEE193 pKa = 4.35SKK195 pKa = 10.43KK196 pKa = 10.53VGSSTSTPNPEE207 pKa = 4.37NITPVTINQDD217 pKa = 3.72NISKK221 pKa = 9.95IFKK224 pKa = 10.13GEE226 pKa = 4.03EE227 pKa = 3.66IGEE230 pKa = 4.26SEE232 pKa = 4.41QLFSTISAYY241 pKa = 9.53TEE243 pKa = 4.01VEE245 pKa = 3.97KK246 pKa = 11.1LLDD249 pKa = 4.57SICSEE254 pKa = 4.35HH255 pKa = 7.0PLDD258 pKa = 4.66SRR260 pKa = 11.84INKK263 pKa = 9.53GKK265 pKa = 10.7FEE267 pKa = 4.02AQIALLDD274 pKa = 3.61TNKK277 pKa = 10.11IIKK280 pKa = 8.56CKK282 pKa = 9.69PMQYY286 pKa = 10.22SPQDD290 pKa = 3.24RR291 pKa = 11.84EE292 pKa = 3.95EE293 pKa = 4.08FKK295 pKa = 10.25TQIEE299 pKa = 4.31EE300 pKa = 4.11LLKK303 pKa = 10.97LGIIRR308 pKa = 11.84PSKK311 pKa = 10.44SPHH314 pKa = 6.05SSPAFMVRR322 pKa = 11.84NHH324 pKa = 6.76AEE326 pKa = 3.83IKK328 pKa = 9.7RR329 pKa = 11.84GKK331 pKa = 10.15ARR333 pKa = 11.84MVINYY338 pKa = 8.06KK339 pKa = 10.32KK340 pKa = 10.96LNDD343 pKa = 3.53HH344 pKa = 6.52TKK346 pKa = 10.7GDD348 pKa = 4.18GYY350 pKa = 11.37LLPNKK355 pKa = 9.54EE356 pKa = 4.03QLLQRR361 pKa = 11.84IGGKK365 pKa = 7.24TFYY368 pKa = 11.14SSFDD372 pKa = 3.75CKK374 pKa = 10.91SGFWQVRR381 pKa = 11.84LAPEE385 pKa = 5.06TIQLTAFSCPQGHH398 pKa = 5.69YY399 pKa = 9.03EE400 pKa = 3.94WLVMPFGLKK409 pKa = 9.04QAPAIFQRR417 pKa = 11.84HH418 pKa = 4.77MDD420 pKa = 3.46EE421 pKa = 4.83SLSNMYY427 pKa = 9.71PQFCAVYY434 pKa = 9.19VDD436 pKa = 4.82DD437 pKa = 6.12IIVFSKK443 pKa = 10.0TEE445 pKa = 4.01EE446 pKa = 3.83EE447 pKa = 4.28HH448 pKa = 6.5LGHH451 pKa = 6.61VKK453 pKa = 10.05IVLNRR458 pKa = 11.84CKK460 pKa = 10.63ALGIVLSKK468 pKa = 10.87KK469 pKa = 9.87KK470 pKa = 10.54AQLCKK475 pKa = 8.61TTINFLGLVIEE486 pKa = 4.63RR487 pKa = 11.84GNLKK491 pKa = 9.61VQSHH495 pKa = 6.75IGLHH499 pKa = 5.94LVAFPDD505 pKa = 3.61QLSDD509 pKa = 3.36RR510 pKa = 11.84NALQRR515 pKa = 11.84FLGLLNYY522 pKa = 9.96ISAYY526 pKa = 9.1FPKK529 pKa = 10.03IANLRR534 pKa = 11.84SPLQVKK540 pKa = 9.46LKK542 pKa = 11.05KK543 pKa = 10.31EE544 pKa = 4.27ITWSWTEE551 pKa = 3.54KK552 pKa = 9.89DD553 pKa = 3.3TEE555 pKa = 4.38TVRR558 pKa = 11.84KK559 pKa = 9.39IKK561 pKa = 10.77SLVKK565 pKa = 9.74TLPDD569 pKa = 4.37LYY571 pKa = 11.08NPSPEE576 pKa = 4.2DD577 pKa = 3.66KK578 pKa = 10.84PIIEE582 pKa = 5.18CDD584 pKa = 3.38ASDD587 pKa = 3.98DD588 pKa = 3.4HH589 pKa = 6.84WGAILKK595 pKa = 10.54AKK597 pKa = 10.21LPEE600 pKa = 4.37GKK602 pKa = 9.94EE603 pKa = 4.14VICRR607 pKa = 11.84YY608 pKa = 10.34ASGTFKK614 pKa = 10.46PAEE617 pKa = 4.16KK618 pKa = 10.04NYY620 pKa = 10.23HH621 pKa = 5.5SNEE624 pKa = 3.89KK625 pKa = 10.26EE626 pKa = 3.54ILSIIKK632 pKa = 9.95AIKK635 pKa = 9.99AFRR638 pKa = 11.84AYY640 pKa = 9.61ILPYY644 pKa = 9.78KK645 pKa = 10.45FLVRR649 pKa = 11.84TDD651 pKa = 3.64NTNAAYY657 pKa = 9.54FVRR660 pKa = 11.84TNIAGDD666 pKa = 3.7YY667 pKa = 8.76KK668 pKa = 10.47QSRR671 pKa = 11.84LVRR674 pKa = 11.84WQMALRR680 pKa = 11.84EE681 pKa = 3.91YY682 pKa = 10.99SFDD685 pKa = 3.45IEE687 pKa = 4.39HH688 pKa = 7.15VSGQKK693 pKa = 10.16NVLADD698 pKa = 3.1IMTRR702 pKa = 11.84EE703 pKa = 4.15LAGKK707 pKa = 7.03TT708 pKa = 3.48
MM1 pKa = 7.66EE2 pKa = 5.88NNLQLLEE9 pKa = 4.22EE10 pKa = 4.68LNHH13 pKa = 7.69LEE15 pKa = 4.73EE16 pKa = 4.64GWEE19 pKa = 3.95MLTDD23 pKa = 3.8EE24 pKa = 5.47DD25 pKa = 6.11FKK27 pKa = 11.43LQQNEE32 pKa = 4.33DD33 pKa = 3.81EE34 pKa = 4.43EE35 pKa = 4.46QLLEE39 pKa = 4.29SVDD42 pKa = 3.62SEE44 pKa = 4.44EE45 pKa = 5.18RR46 pKa = 11.84FSTLTKK52 pKa = 10.01TNPNSIYY59 pKa = 10.23IRR61 pKa = 11.84GNFYY65 pKa = 10.26FKK67 pKa = 10.67GYY69 pKa = 10.34KK70 pKa = 9.7KK71 pKa = 10.74YY72 pKa = 11.06SLDD75 pKa = 3.79LYY77 pKa = 10.59VDD79 pKa = 4.32TGASMCTANKK89 pKa = 10.02HH90 pKa = 5.49VIPEE94 pKa = 4.23EE95 pKa = 3.76FWVNAKK101 pKa = 10.44NPIRR105 pKa = 11.84ARR107 pKa = 11.84IANDD111 pKa = 3.53SIMTFNKK118 pKa = 9.43VAEE121 pKa = 4.13LMQVQIADD129 pKa = 3.66EE130 pKa = 4.3TFIIPTLYY138 pKa = 10.45QATTKK143 pKa = 11.15GDD145 pKa = 3.12ITLGNNFCRR154 pKa = 11.84LYY156 pKa = 10.98EE157 pKa = 4.01PFVQYY162 pKa = 11.36KK163 pKa = 11.03DD164 pKa = 3.49MITFHH169 pKa = 6.96KK170 pKa = 10.33DD171 pKa = 2.48GRR173 pKa = 11.84AVSTKK178 pKa = 10.15KK179 pKa = 8.48VTKK182 pKa = 10.07AYY184 pKa = 10.35FHH186 pKa = 6.73GLPGFLEE193 pKa = 4.35SKK195 pKa = 10.43KK196 pKa = 10.53VGSSTSTPNPEE207 pKa = 4.37NITPVTINQDD217 pKa = 3.72NISKK221 pKa = 9.95IFKK224 pKa = 10.13GEE226 pKa = 4.03EE227 pKa = 3.66IGEE230 pKa = 4.26SEE232 pKa = 4.41QLFSTISAYY241 pKa = 9.53TEE243 pKa = 4.01VEE245 pKa = 3.97KK246 pKa = 11.1LLDD249 pKa = 4.57SICSEE254 pKa = 4.35HH255 pKa = 7.0PLDD258 pKa = 4.66SRR260 pKa = 11.84INKK263 pKa = 9.53GKK265 pKa = 10.7FEE267 pKa = 4.02AQIALLDD274 pKa = 3.61TNKK277 pKa = 10.11IIKK280 pKa = 8.56CKK282 pKa = 9.69PMQYY286 pKa = 10.22SPQDD290 pKa = 3.24RR291 pKa = 11.84EE292 pKa = 3.95EE293 pKa = 4.08FKK295 pKa = 10.25TQIEE299 pKa = 4.31EE300 pKa = 4.11LLKK303 pKa = 10.97LGIIRR308 pKa = 11.84PSKK311 pKa = 10.44SPHH314 pKa = 6.05SSPAFMVRR322 pKa = 11.84NHH324 pKa = 6.76AEE326 pKa = 3.83IKK328 pKa = 9.7RR329 pKa = 11.84GKK331 pKa = 10.15ARR333 pKa = 11.84MVINYY338 pKa = 8.06KK339 pKa = 10.32KK340 pKa = 10.96LNDD343 pKa = 3.53HH344 pKa = 6.52TKK346 pKa = 10.7GDD348 pKa = 4.18GYY350 pKa = 11.37LLPNKK355 pKa = 9.54EE356 pKa = 4.03QLLQRR361 pKa = 11.84IGGKK365 pKa = 7.24TFYY368 pKa = 11.14SSFDD372 pKa = 3.75CKK374 pKa = 10.91SGFWQVRR381 pKa = 11.84LAPEE385 pKa = 5.06TIQLTAFSCPQGHH398 pKa = 5.69YY399 pKa = 9.03EE400 pKa = 3.94WLVMPFGLKK409 pKa = 9.04QAPAIFQRR417 pKa = 11.84HH418 pKa = 4.77MDD420 pKa = 3.46EE421 pKa = 4.83SLSNMYY427 pKa = 9.71PQFCAVYY434 pKa = 9.19VDD436 pKa = 4.82DD437 pKa = 6.12IIVFSKK443 pKa = 10.0TEE445 pKa = 4.01EE446 pKa = 3.83EE447 pKa = 4.28HH448 pKa = 6.5LGHH451 pKa = 6.61VKK453 pKa = 10.05IVLNRR458 pKa = 11.84CKK460 pKa = 10.63ALGIVLSKK468 pKa = 10.87KK469 pKa = 9.87KK470 pKa = 10.54AQLCKK475 pKa = 8.61TTINFLGLVIEE486 pKa = 4.63RR487 pKa = 11.84GNLKK491 pKa = 9.61VQSHH495 pKa = 6.75IGLHH499 pKa = 5.94LVAFPDD505 pKa = 3.61QLSDD509 pKa = 3.36RR510 pKa = 11.84NALQRR515 pKa = 11.84FLGLLNYY522 pKa = 9.96ISAYY526 pKa = 9.1FPKK529 pKa = 10.03IANLRR534 pKa = 11.84SPLQVKK540 pKa = 9.46LKK542 pKa = 11.05KK543 pKa = 10.31EE544 pKa = 4.27ITWSWTEE551 pKa = 3.54KK552 pKa = 9.89DD553 pKa = 3.3TEE555 pKa = 4.38TVRR558 pKa = 11.84KK559 pKa = 9.39IKK561 pKa = 10.77SLVKK565 pKa = 9.74TLPDD569 pKa = 4.37LYY571 pKa = 11.08NPSPEE576 pKa = 4.2DD577 pKa = 3.66KK578 pKa = 10.84PIIEE582 pKa = 5.18CDD584 pKa = 3.38ASDD587 pKa = 3.98DD588 pKa = 3.4HH589 pKa = 6.84WGAILKK595 pKa = 10.54AKK597 pKa = 10.21LPEE600 pKa = 4.37GKK602 pKa = 9.94EE603 pKa = 4.14VICRR607 pKa = 11.84YY608 pKa = 10.34ASGTFKK614 pKa = 10.46PAEE617 pKa = 4.16KK618 pKa = 10.04NYY620 pKa = 10.23HH621 pKa = 5.5SNEE624 pKa = 3.89KK625 pKa = 10.26EE626 pKa = 3.54ILSIIKK632 pKa = 9.95AIKK635 pKa = 9.99AFRR638 pKa = 11.84AYY640 pKa = 9.61ILPYY644 pKa = 9.78KK645 pKa = 10.45FLVRR649 pKa = 11.84TDD651 pKa = 3.64NTNAAYY657 pKa = 9.54FVRR660 pKa = 11.84TNIAGDD666 pKa = 3.7YY667 pKa = 8.76KK668 pKa = 10.47QSRR671 pKa = 11.84LVRR674 pKa = 11.84WQMALRR680 pKa = 11.84EE681 pKa = 3.91YY682 pKa = 10.99SFDD685 pKa = 3.45IEE687 pKa = 4.39HH688 pKa = 7.15VSGQKK693 pKa = 10.16NVLADD698 pKa = 3.1IMTRR702 pKa = 11.84EE703 pKa = 4.15LAGKK707 pKa = 7.03TT708 pKa = 3.48
Molecular weight: 81.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2302 |
117 |
708 |
383.7 |
44.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.778 ± 0.806 | 1.781 ± 0.313 |
5.083 ± 0.25 | 9.296 ± 0.793 |
3.475 ± 0.497 | 4.083 ± 0.453 |
2.433 ± 0.284 | 7.081 ± 0.457 |
9.6 ± 0.516 | 9.513 ± 0.267 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.738 ± 0.282 | 5.039 ± 0.461 |
4.344 ± 0.313 | 4.561 ± 0.669 |
5.213 ± 0.476 | 7.515 ± 0.367 |
5.908 ± 0.117 | 3.736 ± 0.449 |
0.956 ± 0.119 | 3.866 ± 0.338 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
