
Gordonia phage Lozinak
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Smoothievirus; unclassified Smoothievirus
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
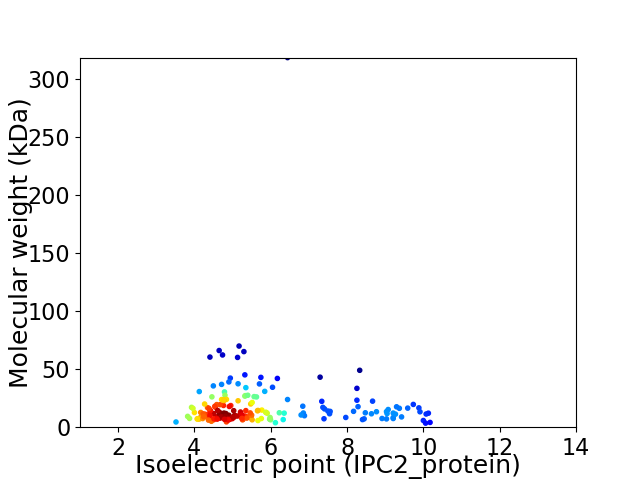
Virtual 2D-PAGE plot for 179 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D1GGA1|A0A2D1GGA1_9CAUD Uncharacterized protein OS=Gordonia phage Lozinak OX=2041513 GN=SEA_LOZINAK_171 PE=4 SV=1
MM1 pKa = 6.93STIDD5 pKa = 3.04ITTQVLAEE13 pKa = 4.16MWTEE17 pKa = 3.46NTGRR21 pKa = 11.84HH22 pKa = 5.21FLDD25 pKa = 3.38SGGAYY30 pKa = 8.98GRR32 pKa = 11.84SWEE35 pKa = 4.14RR36 pKa = 11.84NQAATAGVDD45 pKa = 2.9AGEE48 pKa = 4.53YY49 pKa = 10.13FLSLPQVTLDD59 pKa = 3.2ASYY62 pKa = 10.98NYY64 pKa = 10.46VDD66 pKa = 3.23ITVSAFHH73 pKa = 7.05WLNEE77 pKa = 3.91ILDD80 pKa = 3.82YY81 pKa = 11.08APRR84 pKa = 11.84LNRR87 pKa = 11.84MFEE90 pKa = 4.05MFVALEE96 pKa = 4.25DD97 pKa = 3.76EE98 pKa = 4.84RR99 pKa = 11.84DD100 pKa = 3.98RR101 pKa = 11.84PWMAEE106 pKa = 3.35AEE108 pKa = 4.45AFVEE112 pKa = 4.03WLEE115 pKa = 4.05QHH117 pKa = 6.12GRR119 pKa = 11.84ASDD122 pKa = 3.62VQTVNTYY129 pKa = 10.71NGEE132 pKa = 4.05TWLDD136 pKa = 3.25STLQYY141 pKa = 11.59VVFTYY146 pKa = 9.98TDD148 pKa = 3.34RR149 pKa = 11.84DD150 pKa = 3.81GFDD153 pKa = 3.34TDD155 pKa = 3.79MVMLQYY161 pKa = 10.73HH162 pKa = 6.75GGCDD166 pKa = 3.31VRR168 pKa = 11.84AGYY171 pKa = 7.45TKK173 pKa = 10.35PRR175 pKa = 11.84AFSIGGYY182 pKa = 7.03EE183 pKa = 4.14GRR185 pKa = 11.84YY186 pKa = 8.37EE187 pKa = 4.74LYY189 pKa = 10.22TEE191 pKa = 4.28GHH193 pKa = 5.94VSLSCTEE200 pKa = 4.27NYY202 pKa = 9.56GQEE205 pKa = 3.94IGVGLDD211 pKa = 2.84GDD213 pKa = 4.03TVYY216 pKa = 11.14AGTHH220 pKa = 5.29AWDD223 pKa = 4.11SNSSGVDD230 pKa = 2.73WVEE233 pKa = 3.45YY234 pKa = 10.26GGAYY238 pKa = 7.82DD239 pKa = 3.86TPDD242 pKa = 3.35FEE244 pKa = 5.08TIEE247 pKa = 4.62PEE249 pKa = 4.52DD250 pKa = 4.37DD251 pKa = 3.28GTPYY255 pKa = 10.07VACPTCKK262 pKa = 10.39APLQVSVHH270 pKa = 6.5FGHH273 pKa = 7.73
MM1 pKa = 6.93STIDD5 pKa = 3.04ITTQVLAEE13 pKa = 4.16MWTEE17 pKa = 3.46NTGRR21 pKa = 11.84HH22 pKa = 5.21FLDD25 pKa = 3.38SGGAYY30 pKa = 8.98GRR32 pKa = 11.84SWEE35 pKa = 4.14RR36 pKa = 11.84NQAATAGVDD45 pKa = 2.9AGEE48 pKa = 4.53YY49 pKa = 10.13FLSLPQVTLDD59 pKa = 3.2ASYY62 pKa = 10.98NYY64 pKa = 10.46VDD66 pKa = 3.23ITVSAFHH73 pKa = 7.05WLNEE77 pKa = 3.91ILDD80 pKa = 3.82YY81 pKa = 11.08APRR84 pKa = 11.84LNRR87 pKa = 11.84MFEE90 pKa = 4.05MFVALEE96 pKa = 4.25DD97 pKa = 3.76EE98 pKa = 4.84RR99 pKa = 11.84DD100 pKa = 3.98RR101 pKa = 11.84PWMAEE106 pKa = 3.35AEE108 pKa = 4.45AFVEE112 pKa = 4.03WLEE115 pKa = 4.05QHH117 pKa = 6.12GRR119 pKa = 11.84ASDD122 pKa = 3.62VQTVNTYY129 pKa = 10.71NGEE132 pKa = 4.05TWLDD136 pKa = 3.25STLQYY141 pKa = 11.59VVFTYY146 pKa = 9.98TDD148 pKa = 3.34RR149 pKa = 11.84DD150 pKa = 3.81GFDD153 pKa = 3.34TDD155 pKa = 3.79MVMLQYY161 pKa = 10.73HH162 pKa = 6.75GGCDD166 pKa = 3.31VRR168 pKa = 11.84AGYY171 pKa = 7.45TKK173 pKa = 10.35PRR175 pKa = 11.84AFSIGGYY182 pKa = 7.03EE183 pKa = 4.14GRR185 pKa = 11.84YY186 pKa = 8.37EE187 pKa = 4.74LYY189 pKa = 10.22TEE191 pKa = 4.28GHH193 pKa = 5.94VSLSCTEE200 pKa = 4.27NYY202 pKa = 9.56GQEE205 pKa = 3.94IGVGLDD211 pKa = 2.84GDD213 pKa = 4.03TVYY216 pKa = 11.14AGTHH220 pKa = 5.29AWDD223 pKa = 4.11SNSSGVDD230 pKa = 2.73WVEE233 pKa = 3.45YY234 pKa = 10.26GGAYY238 pKa = 7.82DD239 pKa = 3.86TPDD242 pKa = 3.35FEE244 pKa = 5.08TIEE247 pKa = 4.62PEE249 pKa = 4.52DD250 pKa = 4.37DD251 pKa = 3.28GTPYY255 pKa = 10.07VACPTCKK262 pKa = 10.39APLQVSVHH270 pKa = 6.5FGHH273 pKa = 7.73
Molecular weight: 30.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D1GFV2|A0A2D1GFV2_9CAUD Uncharacterized protein OS=Gordonia phage Lozinak OX=2041513 GN=SEA_LOZINAK_105 PE=4 SV=1
MM1 pKa = 7.39PRR3 pKa = 11.84SRR5 pKa = 11.84SWSDD9 pKa = 3.05DD10 pKa = 3.38DD11 pKa = 4.9LRR13 pKa = 11.84AAVRR17 pKa = 11.84SSQTYY22 pKa = 10.19ADD24 pKa = 3.69VARR27 pKa = 11.84ALSMHH32 pKa = 6.94PGSSTVGRR40 pKa = 11.84LRR42 pKa = 11.84DD43 pKa = 3.46RR44 pKa = 11.84TRR46 pKa = 11.84ALGLDD51 pKa = 3.31TRR53 pKa = 11.84HH54 pKa = 5.64FTRR57 pKa = 11.84ARR59 pKa = 11.84PRR61 pKa = 11.84EE62 pKa = 3.81DD63 pKa = 2.83MFRR66 pKa = 11.84SGVRR70 pKa = 11.84YY71 pKa = 7.38HH72 pKa = 6.2TGMRR76 pKa = 11.84DD77 pKa = 2.94RR78 pKa = 11.84YY79 pKa = 8.64LQLMPYY85 pKa = 8.75EE86 pKa = 4.24CQICGVSKK94 pKa = 10.15WCGQPLTLQLDD105 pKa = 4.43HH106 pKa = 7.59IDD108 pKa = 3.63GDD110 pKa = 4.0RR111 pKa = 11.84MNNTLQNLRR120 pKa = 11.84LLCPNCHH127 pKa = 6.03SQTATWSRR135 pKa = 11.84KK136 pKa = 8.97KK137 pKa = 10.77SNPVV141 pKa = 2.8
MM1 pKa = 7.39PRR3 pKa = 11.84SRR5 pKa = 11.84SWSDD9 pKa = 3.05DD10 pKa = 3.38DD11 pKa = 4.9LRR13 pKa = 11.84AAVRR17 pKa = 11.84SSQTYY22 pKa = 10.19ADD24 pKa = 3.69VARR27 pKa = 11.84ALSMHH32 pKa = 6.94PGSSTVGRR40 pKa = 11.84LRR42 pKa = 11.84DD43 pKa = 3.46RR44 pKa = 11.84TRR46 pKa = 11.84ALGLDD51 pKa = 3.31TRR53 pKa = 11.84HH54 pKa = 5.64FTRR57 pKa = 11.84ARR59 pKa = 11.84PRR61 pKa = 11.84EE62 pKa = 3.81DD63 pKa = 2.83MFRR66 pKa = 11.84SGVRR70 pKa = 11.84YY71 pKa = 7.38HH72 pKa = 6.2TGMRR76 pKa = 11.84DD77 pKa = 2.94RR78 pKa = 11.84YY79 pKa = 8.64LQLMPYY85 pKa = 8.75EE86 pKa = 4.24CQICGVSKK94 pKa = 10.15WCGQPLTLQLDD105 pKa = 4.43HH106 pKa = 7.59IDD108 pKa = 3.63GDD110 pKa = 4.0RR111 pKa = 11.84MNNTLQNLRR120 pKa = 11.84LLCPNCHH127 pKa = 6.03SQTATWSRR135 pKa = 11.84KK136 pKa = 8.97KK137 pKa = 10.77SNPVV141 pKa = 2.8
Molecular weight: 16.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
29354 |
27 |
2931 |
164.0 |
18.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.576 ± 0.33 | 1.036 ± 0.126 |
7.137 ± 0.182 | 6.752 ± 0.206 |
3.158 ± 0.111 | 7.856 ± 0.241 |
2.34 ± 0.146 | 4.735 ± 0.155 |
4.085 ± 0.169 | 7.624 ± 0.168 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.501 ± 0.099 | 3.655 ± 0.149 |
4.889 ± 0.173 | 3.655 ± 0.217 |
6.984 ± 0.209 | 5.73 ± 0.146 |
5.86 ± 0.188 | 7.375 ± 0.269 |
2.146 ± 0.095 | 2.906 ± 0.145 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
