
Monodon monoceros (Narwhal) (Ceratodon monodon)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria; Boreoeutheria; Laurasiatheria; Artiodactyla; Whippomorpha;
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
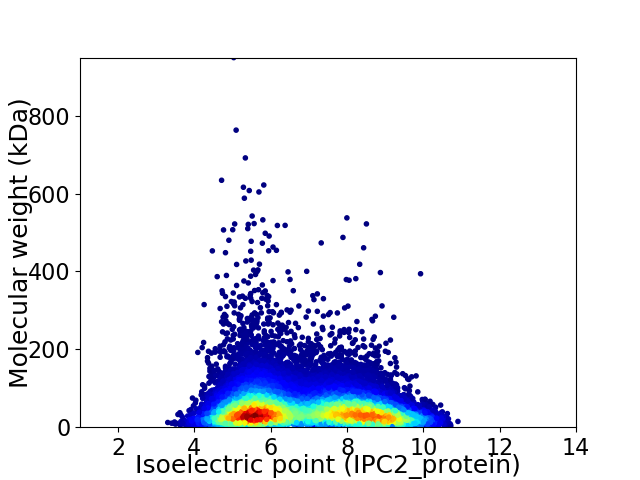
Virtual 2D-PAGE plot for 20731 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4U1F987|A0A4U1F987_MONMO LRRCT domain-containing protein (Fragment) OS=Monodon monoceros OX=40151 GN=EI555_005442 PE=4 SV=1
MM1 pKa = 7.58PADD4 pKa = 3.72PAATKK9 pKa = 10.64APGMEE14 pKa = 4.23SSGSVAGLGEE24 pKa = 4.1VDD26 pKa = 5.37LGTFLDD32 pKa = 4.85KK33 pKa = 11.15DD34 pKa = 4.12SEE36 pKa = 4.38CDD38 pKa = 3.39SGIEE42 pKa = 3.93VCVCNWGHH50 pKa = 6.92HH51 pKa = 7.02DD52 pKa = 5.35DD53 pKa = 4.8DD54 pKa = 4.67LKK56 pKa = 11.28EE57 pKa = 4.17FNVLIDD63 pKa = 4.67DD64 pKa = 4.61ALDD67 pKa = 3.69MPLDD71 pKa = 4.1FCDD74 pKa = 3.73SCHH77 pKa = 6.31VLPPGDD83 pKa = 3.84EE84 pKa = 4.28EE85 pKa = 4.57EE86 pKa = 5.22GLGQPSEE93 pKa = 4.39GCSWPSASAPGGVHH107 pKa = 5.86NRR109 pKa = 11.84SGQKK113 pKa = 10.53AKK115 pKa = 10.9VEE117 pKa = 4.14PQTEE121 pKa = 4.14AGLDD125 pKa = 3.71LQCQARR131 pKa = 11.84TGCGPDD137 pKa = 3.21VSPAQNCQEE146 pKa = 4.11GLVATTVLL154 pKa = 3.71
MM1 pKa = 7.58PADD4 pKa = 3.72PAATKK9 pKa = 10.64APGMEE14 pKa = 4.23SSGSVAGLGEE24 pKa = 4.1VDD26 pKa = 5.37LGTFLDD32 pKa = 4.85KK33 pKa = 11.15DD34 pKa = 4.12SEE36 pKa = 4.38CDD38 pKa = 3.39SGIEE42 pKa = 3.93VCVCNWGHH50 pKa = 6.92HH51 pKa = 7.02DD52 pKa = 5.35DD53 pKa = 4.8DD54 pKa = 4.67LKK56 pKa = 11.28EE57 pKa = 4.17FNVLIDD63 pKa = 4.67DD64 pKa = 4.61ALDD67 pKa = 3.69MPLDD71 pKa = 4.1FCDD74 pKa = 3.73SCHH77 pKa = 6.31VLPPGDD83 pKa = 3.84EE84 pKa = 4.28EE85 pKa = 4.57EE86 pKa = 5.22GLGQPSEE93 pKa = 4.39GCSWPSASAPGGVHH107 pKa = 5.86NRR109 pKa = 11.84SGQKK113 pKa = 10.53AKK115 pKa = 10.9VEE117 pKa = 4.14PQTEE121 pKa = 4.14AGLDD125 pKa = 3.71LQCQARR131 pKa = 11.84TGCGPDD137 pKa = 3.21VSPAQNCQEE146 pKa = 4.11GLVATTVLL154 pKa = 3.71
Molecular weight: 15.96 kDa
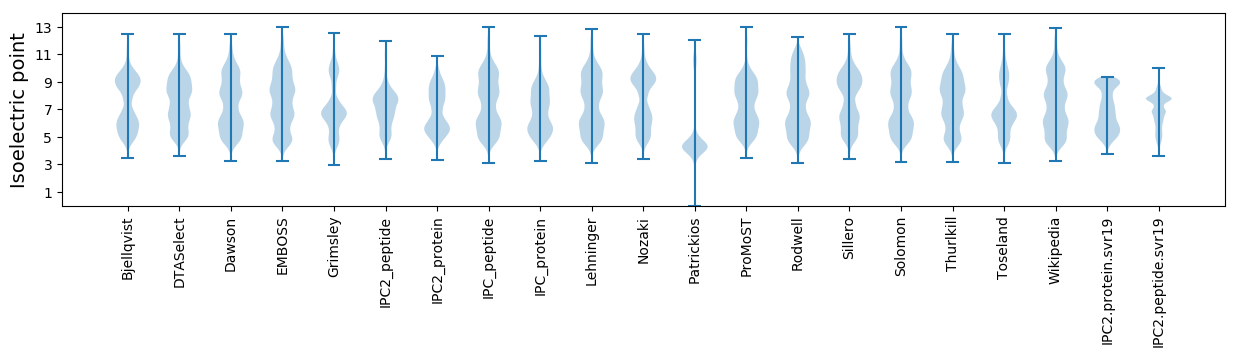
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4U1FQ66|A0A4U1FQ66_MONMO Deacetylase sirtuin-type domain-containing protein (Fragment) OS=Monodon monoceros OX=40151 GN=EI555_021280 PE=3 SV=1
PP1 pKa = 7.13FLPWGAGSRR10 pKa = 11.84GDD12 pKa = 3.55LGGWSWLGAASALFEE27 pKa = 4.61PRR29 pKa = 11.84SDD31 pKa = 3.98PDD33 pKa = 3.94LPPDD37 pKa = 3.72NPAPLAEE44 pKa = 4.26LQQPLSGSAPNPQMPLGWLSPTSHH68 pKa = 6.95IAQFRR73 pKa = 11.84RR74 pKa = 11.84GDD76 pKa = 3.88PVPASALGLGLLLSRR91 pKa = 11.84ITRR94 pKa = 11.84TKK96 pKa = 10.55LGSLSFGGWGRR107 pKa = 11.84PGRR110 pKa = 11.84RR111 pKa = 11.84SPEE114 pKa = 3.57RR115 pKa = 11.84RR116 pKa = 11.84GEE118 pKa = 3.94QGVAAGGAPGPPNRR132 pKa = 11.84AGDD135 pKa = 3.63EE136 pKa = 4.07PARR139 pKa = 11.84GGGPAAASPRR149 pKa = 11.84LAARR153 pKa = 11.84EE154 pKa = 3.8RR155 pKa = 11.84VGRR158 pKa = 11.84GGGGEE163 pKa = 4.01PAGLLAGGRR172 pKa = 11.84AGSPEE177 pKa = 3.53PRR179 pKa = 11.84EE180 pKa = 4.16RR181 pKa = 11.84RR182 pKa = 11.84GSSAPSLPRR191 pKa = 11.84SSRR194 pKa = 11.84VSPFHH199 pKa = 6.99PDD201 pKa = 2.73ARR203 pKa = 11.84PAGLLHH209 pKa = 6.72GSALRR214 pKa = 11.84AQEE217 pKa = 4.06AKK219 pKa = 10.49AGEE222 pKa = 4.54AMCQPGWRR230 pKa = 11.84APLGVPRR237 pKa = 11.84TSPGAATAAAAAAAAAAAAAQPALASPGEE266 pKa = 4.57GATAPRR272 pKa = 11.84DD273 pKa = 4.01FPNPAYY279 pKa = 10.07HH280 pKa = 6.65GHH282 pKa = 6.34WFWASLRR289 pKa = 11.84LLRR292 pKa = 11.84SPRR295 pKa = 11.84SSSEE299 pKa = 3.96LEE301 pKa = 4.09NFSCVPQTHH310 pKa = 7.21LAQQRR315 pKa = 11.84VLQDD319 pKa = 3.35AGSASPASRR328 pKa = 11.84KK329 pKa = 6.29TTGHH333 pKa = 6.53CPQFPPNLGIVPGSSNGLGVAPGHH357 pKa = 6.58WEE359 pKa = 3.98LPPGSRR365 pKa = 11.84SPSARR370 pKa = 11.84PPPVAEE376 pKa = 4.2RR377 pKa = 11.84SLSVSAPSAPADD389 pKa = 3.47AASGLKK395 pKa = 9.63PRR397 pKa = 11.84AAAGDD402 pKa = 3.66TGAGRR407 pKa = 11.84GSLRR411 pKa = 11.84RR412 pKa = 11.84ASPSAAALARR422 pKa = 11.84PGALKK427 pKa = 9.67PAPLSFALCKK437 pKa = 10.59DD438 pKa = 3.25HH439 pKa = 6.74GHH441 pKa = 6.1PQIIVVAPGAEE452 pKa = 4.1PEE454 pKa = 4.32STQVQRR460 pKa = 11.84TEE462 pKa = 4.04DD463 pKa = 3.41GGTIIRR469 pKa = 11.84IFWVGPKK476 pKa = 10.58GEE478 pKa = 4.81LLRR481 pKa = 11.84CTPVSPVMQTPMGIRR496 pKa = 11.84VGKK499 pKa = 10.78SMLGLLAFLAFASCCYY515 pKa = 10.05AAYY518 pKa = 10.06RR519 pKa = 11.84PSEE522 pKa = 4.16TLCGGEE528 pKa = 5.39LVDD531 pKa = 4.66TLQFVCGDD539 pKa = 3.1RR540 pKa = 11.84GFYY543 pKa = 10.25FSRR546 pKa = 11.84PASRR550 pKa = 11.84VNRR553 pKa = 11.84RR554 pKa = 11.84SRR556 pKa = 11.84GIVEE560 pKa = 4.06EE561 pKa = 4.48CCFRR565 pKa = 11.84SCDD568 pKa = 3.56LALLEE573 pKa = 5.08TYY575 pKa = 10.26CATPAKK581 pKa = 10.1SEE583 pKa = 4.34RR584 pKa = 11.84DD585 pKa = 3.4VSTPPTVLPDD595 pKa = 3.31NFPRR599 pKa = 11.84YY600 pKa = 8.84PVGKK604 pKa = 9.78FFQYY608 pKa = 9.48DD609 pKa = 3.09TWKK612 pKa = 10.71QSAQRR617 pKa = 11.84LRR619 pKa = 11.84RR620 pKa = 11.84GLPALLRR627 pKa = 11.84ARR629 pKa = 11.84RR630 pKa = 11.84GRR632 pKa = 11.84TLAKK636 pKa = 8.82EE637 pKa = 3.92LEE639 pKa = 4.44VFRR642 pKa = 11.84EE643 pKa = 4.6AKK645 pKa = 9.39RR646 pKa = 11.84HH647 pKa = 5.57RR648 pKa = 11.84PLIALPTQDD657 pKa = 3.74PAAHH661 pKa = 6.65GGASLEE667 pKa = 4.35ASGHH671 pKa = 4.72RR672 pKa = 3.89
PP1 pKa = 7.13FLPWGAGSRR10 pKa = 11.84GDD12 pKa = 3.55LGGWSWLGAASALFEE27 pKa = 4.61PRR29 pKa = 11.84SDD31 pKa = 3.98PDD33 pKa = 3.94LPPDD37 pKa = 3.72NPAPLAEE44 pKa = 4.26LQQPLSGSAPNPQMPLGWLSPTSHH68 pKa = 6.95IAQFRR73 pKa = 11.84RR74 pKa = 11.84GDD76 pKa = 3.88PVPASALGLGLLLSRR91 pKa = 11.84ITRR94 pKa = 11.84TKK96 pKa = 10.55LGSLSFGGWGRR107 pKa = 11.84PGRR110 pKa = 11.84RR111 pKa = 11.84SPEE114 pKa = 3.57RR115 pKa = 11.84RR116 pKa = 11.84GEE118 pKa = 3.94QGVAAGGAPGPPNRR132 pKa = 11.84AGDD135 pKa = 3.63EE136 pKa = 4.07PARR139 pKa = 11.84GGGPAAASPRR149 pKa = 11.84LAARR153 pKa = 11.84EE154 pKa = 3.8RR155 pKa = 11.84VGRR158 pKa = 11.84GGGGEE163 pKa = 4.01PAGLLAGGRR172 pKa = 11.84AGSPEE177 pKa = 3.53PRR179 pKa = 11.84EE180 pKa = 4.16RR181 pKa = 11.84RR182 pKa = 11.84GSSAPSLPRR191 pKa = 11.84SSRR194 pKa = 11.84VSPFHH199 pKa = 6.99PDD201 pKa = 2.73ARR203 pKa = 11.84PAGLLHH209 pKa = 6.72GSALRR214 pKa = 11.84AQEE217 pKa = 4.06AKK219 pKa = 10.49AGEE222 pKa = 4.54AMCQPGWRR230 pKa = 11.84APLGVPRR237 pKa = 11.84TSPGAATAAAAAAAAAAAAAQPALASPGEE266 pKa = 4.57GATAPRR272 pKa = 11.84DD273 pKa = 4.01FPNPAYY279 pKa = 10.07HH280 pKa = 6.65GHH282 pKa = 6.34WFWASLRR289 pKa = 11.84LLRR292 pKa = 11.84SPRR295 pKa = 11.84SSSEE299 pKa = 3.96LEE301 pKa = 4.09NFSCVPQTHH310 pKa = 7.21LAQQRR315 pKa = 11.84VLQDD319 pKa = 3.35AGSASPASRR328 pKa = 11.84KK329 pKa = 6.29TTGHH333 pKa = 6.53CPQFPPNLGIVPGSSNGLGVAPGHH357 pKa = 6.58WEE359 pKa = 3.98LPPGSRR365 pKa = 11.84SPSARR370 pKa = 11.84PPPVAEE376 pKa = 4.2RR377 pKa = 11.84SLSVSAPSAPADD389 pKa = 3.47AASGLKK395 pKa = 9.63PRR397 pKa = 11.84AAAGDD402 pKa = 3.66TGAGRR407 pKa = 11.84GSLRR411 pKa = 11.84RR412 pKa = 11.84ASPSAAALARR422 pKa = 11.84PGALKK427 pKa = 9.67PAPLSFALCKK437 pKa = 10.59DD438 pKa = 3.25HH439 pKa = 6.74GHH441 pKa = 6.1PQIIVVAPGAEE452 pKa = 4.1PEE454 pKa = 4.32STQVQRR460 pKa = 11.84TEE462 pKa = 4.04DD463 pKa = 3.41GGTIIRR469 pKa = 11.84IFWVGPKK476 pKa = 10.58GEE478 pKa = 4.81LLRR481 pKa = 11.84CTPVSPVMQTPMGIRR496 pKa = 11.84VGKK499 pKa = 10.78SMLGLLAFLAFASCCYY515 pKa = 10.05AAYY518 pKa = 10.06RR519 pKa = 11.84PSEE522 pKa = 4.16TLCGGEE528 pKa = 5.39LVDD531 pKa = 4.66TLQFVCGDD539 pKa = 3.1RR540 pKa = 11.84GFYY543 pKa = 10.25FSRR546 pKa = 11.84PASRR550 pKa = 11.84VNRR553 pKa = 11.84RR554 pKa = 11.84SRR556 pKa = 11.84GIVEE560 pKa = 4.06EE561 pKa = 4.48CCFRR565 pKa = 11.84SCDD568 pKa = 3.56LALLEE573 pKa = 5.08TYY575 pKa = 10.26CATPAKK581 pKa = 10.1SEE583 pKa = 4.34RR584 pKa = 11.84DD585 pKa = 3.4VSTPPTVLPDD595 pKa = 3.31NFPRR599 pKa = 11.84YY600 pKa = 8.84PVGKK604 pKa = 9.78FFQYY608 pKa = 9.48DD609 pKa = 3.09TWKK612 pKa = 10.71QSAQRR617 pKa = 11.84LRR619 pKa = 11.84RR620 pKa = 11.84GLPALLRR627 pKa = 11.84ARR629 pKa = 11.84RR630 pKa = 11.84GRR632 pKa = 11.84TLAKK636 pKa = 8.82EE637 pKa = 3.92LEE639 pKa = 4.44VFRR642 pKa = 11.84EE643 pKa = 4.6AKK645 pKa = 9.39RR646 pKa = 11.84HH647 pKa = 5.57RR648 pKa = 11.84PLIALPTQDD657 pKa = 3.74PAAHH661 pKa = 6.65GGASLEE667 pKa = 4.35ASGHH671 pKa = 4.72RR672 pKa = 3.89
Molecular weight: 70.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
9821986 |
8 |
8564 |
473.8 |
52.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.412 ± 0.021 | 2.255 ± 0.014 |
4.683 ± 0.013 | 6.992 ± 0.021 |
3.567 ± 0.011 | 6.928 ± 0.025 |
2.627 ± 0.01 | 4.094 ± 0.017 |
5.572 ± 0.024 | 9.965 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.02 ± 0.008 | 3.381 ± 0.014 |
6.567 ± 0.03 | 4.687 ± 0.015 |
6.032 ± 0.018 | 8.184 ± 0.021 |
5.21 ± 0.013 | 6.027 ± 0.012 |
1.247 ± 0.006 | 2.534 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
