
Aminobacterium colombiense (strain DSM 12261 / ALA-1)
Taxonomy: cellular organisms; Bacteria; Synergistetes; Synergistia; Synergistales; Synergistaceae; Aminobacterium; Aminobacterium colombiense
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
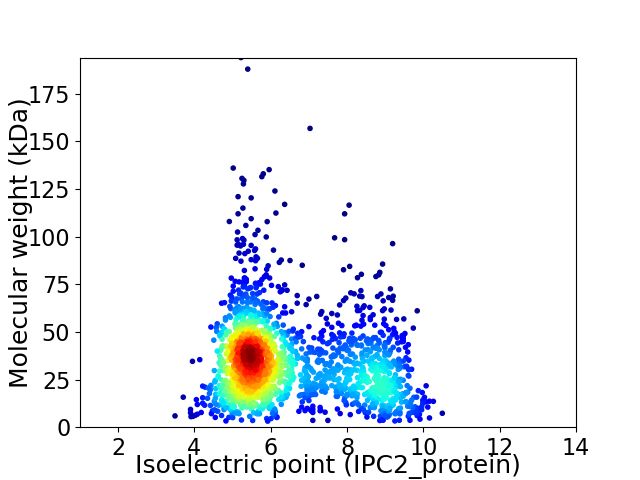
Virtual 2D-PAGE plot for 1872 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D5EFB4|D5EFB4_AMICL Uncharacterized protein OS=Aminobacterium colombiense (strain DSM 12261 / ALA-1) OX=572547 GN=Amico_1122 PE=4 SV=1
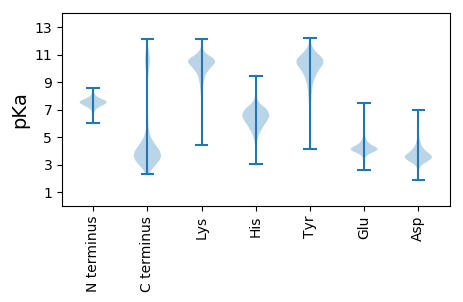
MM1 pKa = 7.26SAKK4 pKa = 10.24EE5 pKa = 3.73KK6 pKa = 10.28IAYY9 pKa = 9.7LKK11 pKa = 11.03GLLAGLNMKK20 pKa = 10.74DD21 pKa = 3.04EE22 pKa = 4.47TAAKK26 pKa = 9.81AFSSVTEE33 pKa = 4.01ALEE36 pKa = 3.99ALADD40 pKa = 4.53EE41 pKa = 4.9IEE43 pKa = 4.16EE44 pKa = 3.94HH45 pKa = 6.4AALIEE50 pKa = 4.13EE51 pKa = 4.22QQDD54 pKa = 3.82LYY56 pKa = 11.61EE57 pKa = 4.21EE58 pKa = 4.95LEE60 pKa = 4.56DD61 pKa = 5.67DD62 pKa = 5.71FSLLDD67 pKa = 3.85EE68 pKa = 5.59DD69 pKa = 5.99LDD71 pKa = 3.85TLEE74 pKa = 4.76RR75 pKa = 11.84SFAEE79 pKa = 4.53FMGEE83 pKa = 4.06DD84 pKa = 3.64FEE86 pKa = 4.68EE87 pKa = 4.35TDD89 pKa = 3.44EE90 pKa = 4.53EE91 pKa = 4.33EE92 pKa = 4.57NEE94 pKa = 3.63FDD96 pKa = 3.39EE97 pKa = 5.92AYY99 pKa = 10.46EE100 pKa = 4.24SVACPKK106 pKa = 9.92CGHH109 pKa = 5.43VFYY112 pKa = 9.97YY113 pKa = 9.44QPEE116 pKa = 4.23MYY118 pKa = 10.24EE119 pKa = 3.85EE120 pKa = 5.48GEE122 pKa = 4.08ALQCPDD128 pKa = 3.99CGEE131 pKa = 4.41TFAQPAEE138 pKa = 4.24EE139 pKa = 4.01
MM1 pKa = 7.26SAKK4 pKa = 10.24EE5 pKa = 3.73KK6 pKa = 10.28IAYY9 pKa = 9.7LKK11 pKa = 11.03GLLAGLNMKK20 pKa = 10.74DD21 pKa = 3.04EE22 pKa = 4.47TAAKK26 pKa = 9.81AFSSVTEE33 pKa = 4.01ALEE36 pKa = 3.99ALADD40 pKa = 4.53EE41 pKa = 4.9IEE43 pKa = 4.16EE44 pKa = 3.94HH45 pKa = 6.4AALIEE50 pKa = 4.13EE51 pKa = 4.22QQDD54 pKa = 3.82LYY56 pKa = 11.61EE57 pKa = 4.21EE58 pKa = 4.95LEE60 pKa = 4.56DD61 pKa = 5.67DD62 pKa = 5.71FSLLDD67 pKa = 3.85EE68 pKa = 5.59DD69 pKa = 5.99LDD71 pKa = 3.85TLEE74 pKa = 4.76RR75 pKa = 11.84SFAEE79 pKa = 4.53FMGEE83 pKa = 4.06DD84 pKa = 3.64FEE86 pKa = 4.68EE87 pKa = 4.35TDD89 pKa = 3.44EE90 pKa = 4.53EE91 pKa = 4.33EE92 pKa = 4.57NEE94 pKa = 3.63FDD96 pKa = 3.39EE97 pKa = 5.92AYY99 pKa = 10.46EE100 pKa = 4.24SVACPKK106 pKa = 9.92CGHH109 pKa = 5.43VFYY112 pKa = 9.97YY113 pKa = 9.44QPEE116 pKa = 4.23MYY118 pKa = 10.24EE119 pKa = 3.85EE120 pKa = 5.48GEE122 pKa = 4.08ALQCPDD128 pKa = 3.99CGEE131 pKa = 4.41TFAQPAEE138 pKa = 4.24EE139 pKa = 4.01
Molecular weight: 15.75 kDa
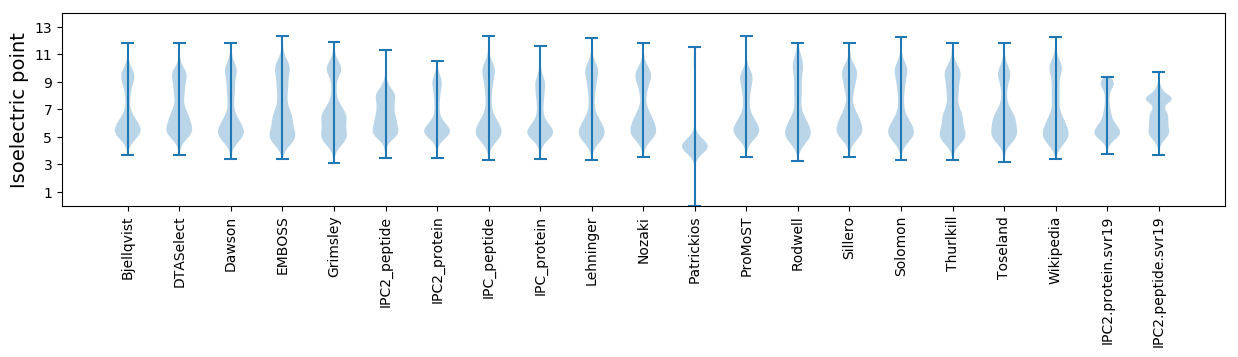
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D5EG22|D5EG22_AMICL Epoxyqueuosine reductase QueH OS=Aminobacterium colombiense (strain DSM 12261 / ALA-1) OX=572547 GN=queH PE=3 SV=1
MM1 pKa = 7.03TAPGRR6 pKa = 11.84MPALFSLAGICLALLVLQQGIPLFIFPLVAMSGTLALVLLFTTQEE51 pKa = 3.93KK52 pKa = 8.8PGYY55 pKa = 7.9WKK57 pKa = 10.81GVTLVVLVAGVGSAYY72 pKa = 9.71ISSRR76 pKa = 11.84LSTDD80 pKa = 3.57FLHH83 pKa = 7.18PGEE86 pKa = 4.38TFQGQGYY93 pKa = 9.65ILLEE97 pKa = 4.33RR98 pKa = 11.84PWGRR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84ALLIRR109 pKa = 11.84TKK111 pKa = 10.63KK112 pKa = 9.5GTFLAKK118 pKa = 9.99IPSQHH123 pKa = 5.82GFIAGDD129 pKa = 3.7TVEE132 pKa = 4.29VAGTMLSLPSSKK144 pKa = 10.19KK145 pKa = 9.79SRR147 pKa = 11.84SFHH150 pKa = 6.26EE151 pKa = 3.8DD152 pKa = 3.43RR153 pKa = 11.84YY154 pKa = 8.72WKK156 pKa = 10.67ARR158 pKa = 11.84GAMAEE163 pKa = 3.92IVIGKK168 pKa = 7.88VEE170 pKa = 4.47KK171 pKa = 10.5IEE173 pKa = 4.22SCRR176 pKa = 11.84QFSLSGWRR184 pKa = 11.84NMLVKK189 pKa = 10.47RR190 pKa = 11.84ILLTQPPRR198 pKa = 11.84TRR200 pKa = 11.84GYY202 pKa = 10.52LAAAWTGMKK211 pKa = 10.25SPLLSSQHH219 pKa = 5.4EE220 pKa = 4.27RR221 pKa = 11.84WGTSHH226 pKa = 7.42LLAVSGFHH234 pKa = 6.57VSLFVSGLLFLLRR247 pKa = 11.84SCLGRR252 pKa = 11.84GFIISWAMWGYY263 pKa = 10.89VLIAGAAPSALRR275 pKa = 11.84AALMMQIALIGRR287 pKa = 11.84CRR289 pKa = 11.84GRR291 pKa = 11.84PSQAVNTVSLAGMALLFWRR310 pKa = 11.84PWWFWDD316 pKa = 4.85LGWQLSMVAALIIAAALEE334 pKa = 4.58RR335 pKa = 11.84GLSPVWWLSVGPAIWVTTLPLAAPIFGDD363 pKa = 3.68VPLAGIVTNLSAGPVFGVLYY383 pKa = 9.92PLASFLALPSLLALPWGNWASQGAEE408 pKa = 3.7GLFILWEE415 pKa = 4.12TWADD419 pKa = 3.77SISRR423 pKa = 11.84LIPVYY428 pKa = 10.73VPWTPFWTAFAAGLLPLLIARR449 pKa = 11.84SLRR452 pKa = 11.84LSWRR456 pKa = 11.84HH457 pKa = 5.47SLAASLLLQCVMLWIVMM474 pKa = 4.14
MM1 pKa = 7.03TAPGRR6 pKa = 11.84MPALFSLAGICLALLVLQQGIPLFIFPLVAMSGTLALVLLFTTQEE51 pKa = 3.93KK52 pKa = 8.8PGYY55 pKa = 7.9WKK57 pKa = 10.81GVTLVVLVAGVGSAYY72 pKa = 9.71ISSRR76 pKa = 11.84LSTDD80 pKa = 3.57FLHH83 pKa = 7.18PGEE86 pKa = 4.38TFQGQGYY93 pKa = 9.65ILLEE97 pKa = 4.33RR98 pKa = 11.84PWGRR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84ALLIRR109 pKa = 11.84TKK111 pKa = 10.63KK112 pKa = 9.5GTFLAKK118 pKa = 9.99IPSQHH123 pKa = 5.82GFIAGDD129 pKa = 3.7TVEE132 pKa = 4.29VAGTMLSLPSSKK144 pKa = 10.19KK145 pKa = 9.79SRR147 pKa = 11.84SFHH150 pKa = 6.26EE151 pKa = 3.8DD152 pKa = 3.43RR153 pKa = 11.84YY154 pKa = 8.72WKK156 pKa = 10.67ARR158 pKa = 11.84GAMAEE163 pKa = 3.92IVIGKK168 pKa = 7.88VEE170 pKa = 4.47KK171 pKa = 10.5IEE173 pKa = 4.22SCRR176 pKa = 11.84QFSLSGWRR184 pKa = 11.84NMLVKK189 pKa = 10.47RR190 pKa = 11.84ILLTQPPRR198 pKa = 11.84TRR200 pKa = 11.84GYY202 pKa = 10.52LAAAWTGMKK211 pKa = 10.25SPLLSSQHH219 pKa = 5.4EE220 pKa = 4.27RR221 pKa = 11.84WGTSHH226 pKa = 7.42LLAVSGFHH234 pKa = 6.57VSLFVSGLLFLLRR247 pKa = 11.84SCLGRR252 pKa = 11.84GFIISWAMWGYY263 pKa = 10.89VLIAGAAPSALRR275 pKa = 11.84AALMMQIALIGRR287 pKa = 11.84CRR289 pKa = 11.84GRR291 pKa = 11.84PSQAVNTVSLAGMALLFWRR310 pKa = 11.84PWWFWDD316 pKa = 4.85LGWQLSMVAALIIAAALEE334 pKa = 4.58RR335 pKa = 11.84GLSPVWWLSVGPAIWVTTLPLAAPIFGDD363 pKa = 3.68VPLAGIVTNLSAGPVFGVLYY383 pKa = 9.92PLASFLALPSLLALPWGNWASQGAEE408 pKa = 3.7GLFILWEE415 pKa = 4.12TWADD419 pKa = 3.77SISRR423 pKa = 11.84LIPVYY428 pKa = 10.73VPWTPFWTAFAAGLLPLLIARR449 pKa = 11.84SLRR452 pKa = 11.84LSWRR456 pKa = 11.84HH457 pKa = 5.47SLAASLLLQCVMLWIVMM474 pKa = 4.14
Molecular weight: 51.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
596225 |
31 |
1759 |
318.5 |
35.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.204 ± 0.051 | 1.227 ± 0.021 |
4.763 ± 0.041 | 7.142 ± 0.063 |
4.3 ± 0.04 | 7.837 ± 0.042 |
1.933 ± 0.026 | 7.109 ± 0.047 |
5.722 ± 0.048 | 10.409 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.77 ± 0.024 | 3.321 ± 0.028 |
4.307 ± 0.034 | 2.94 ± 0.027 |
5.325 ± 0.046 | 6.249 ± 0.039 |
4.799 ± 0.031 | 7.447 ± 0.056 |
1.278 ± 0.023 | 2.918 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
