
Aureicoccus marinus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Aureicoccus
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
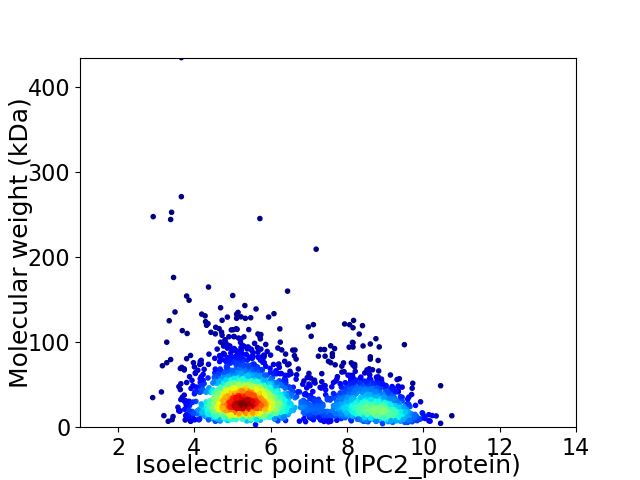
Virtual 2D-PAGE plot for 2543 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S7T6K6|A0A2S7T6K6_9FLAO dTTP/UTP pyrophosphatase OS=Aureicoccus marinus OX=754435 GN=BST99_04785 PE=3 SV=1
MM1 pKa = 7.13NQQFLNPIARR11 pKa = 11.84SMVQVVKK18 pKa = 10.94KK19 pKa = 10.63NLLLYY24 pKa = 10.8GLLLAFGFYY33 pKa = 9.49QANAQVNFDD42 pKa = 3.51QSTLNFNGIGEE53 pKa = 4.47STNGFTSIMFGPDD66 pKa = 2.38GRR68 pKa = 11.84LYY70 pKa = 9.2VAEE73 pKa = 3.86YY74 pKa = 10.11HH75 pKa = 6.23GLIKK79 pKa = 10.41IFTIEE84 pKa = 4.17RR85 pKa = 11.84QSSTSYY91 pKa = 9.94TVTAVEE97 pKa = 4.5EE98 pKa = 4.23LNGIQTMEE106 pKa = 4.31DD107 pKa = 3.6HH108 pKa = 7.05NDD110 pKa = 3.61DD111 pKa = 3.7GSGFSSLEE119 pKa = 3.91RR120 pKa = 11.84EE121 pKa = 4.47TTGLTVAGTATNPIIYY137 pKa = 8.21VTSSDD142 pKa = 3.51FRR144 pKa = 11.84IGSGPAGGNGDD155 pKa = 3.66VGLDD159 pKa = 3.63TNSGVITRR167 pKa = 11.84FTWTGSAWDD176 pKa = 3.55VVDD179 pKa = 5.28LVRR182 pKa = 11.84GLPRR186 pKa = 11.84SEE188 pKa = 4.39EE189 pKa = 3.77NHH191 pKa = 5.43ATNGLEE197 pKa = 3.86YY198 pKa = 10.89AQINGVNYY206 pKa = 10.59LLVTSGGITNAGSPSTNFVFTCEE229 pKa = 4.04YY230 pKa = 9.98ALSGAILSVNLDD242 pKa = 4.42EE243 pKa = 5.82INSLPILTDD252 pKa = 2.84SGRR255 pKa = 11.84QYY257 pKa = 10.9IYY259 pKa = 10.74DD260 pKa = 4.93LPTLDD265 pKa = 5.62DD266 pKa = 3.64PTRR269 pKa = 11.84PNSNGIIDD277 pKa = 4.57PNDD280 pKa = 3.68PGYY283 pKa = 9.57TGEE286 pKa = 5.3DD287 pKa = 3.83LNDD290 pKa = 3.26PWGGNDD296 pKa = 4.14GLNMAIIDD304 pKa = 3.85PAGPVQVFSPGFRR317 pKa = 11.84NAYY320 pKa = 9.45DD321 pKa = 3.63LVVTEE326 pKa = 4.77SGAVYY331 pKa = 8.72ATDD334 pKa = 3.32NGANGGWGGFPVNEE348 pKa = 4.16GTVNANNDD356 pKa = 3.48YY357 pKa = 11.2DD358 pKa = 4.18PNEE361 pKa = 4.4TGSTFATQAPDD372 pKa = 3.36GEE374 pKa = 4.86RR375 pKa = 11.84ILNQDD380 pKa = 3.52HH381 pKa = 7.1LEE383 pKa = 4.68LITTDD388 pKa = 3.42LSSYY392 pKa = 10.47TFGSYY397 pKa = 9.89YY398 pKa = 10.44AGHH401 pKa = 7.38PNPVRR406 pKa = 11.84ANPSGAGLYY415 pKa = 8.17TVNDD419 pKa = 4.13TNSVWRR425 pKa = 11.84TLIYY429 pKa = 10.86DD430 pKa = 4.09PDD432 pKa = 4.16GSSPNSTTDD441 pKa = 3.01VTLGLPANWPPVNTANPDD459 pKa = 3.31EE460 pKa = 4.74GDD462 pKa = 3.01WRR464 pKa = 11.84GPTTPNPDD472 pKa = 3.43GPDD475 pKa = 3.46NSPVLVWGTNTNGIDD490 pKa = 3.69EE491 pKa = 4.38YY492 pKa = 10.69TATNFAGQMQGNLIAGVNSGVLRR515 pKa = 11.84RR516 pKa = 11.84VEE518 pKa = 4.3LNPDD522 pKa = 2.92GSVANFTAEE531 pKa = 4.11FFSGLGGNALAITCNSDD548 pKa = 3.29TEE550 pKa = 4.42IFPGTIWAGTLGGDD564 pKa = 4.53LIVFEE569 pKa = 4.6PQDD572 pKa = 3.81FVVCILPGEE581 pKa = 4.17MDD583 pKa = 4.32YY584 pKa = 11.52DD585 pKa = 4.0PSADD589 pKa = 3.86NDD591 pKa = 3.81SDD593 pKa = 5.24GYY595 pKa = 9.96TNDD598 pKa = 5.87DD599 pKa = 3.58EE600 pKa = 5.03DD601 pKa = 4.75QNGTDD606 pKa = 3.61LCNGGSQPSDD616 pKa = 3.14FDD618 pKa = 3.68KK619 pKa = 11.25AAGGILVSDD628 pKa = 5.05LLDD631 pKa = 4.07NDD633 pKa = 4.93DD634 pKa = 5.07DD635 pKa = 4.94FDD637 pKa = 6.86GIVDD641 pKa = 3.92SADD644 pKa = 3.37PFQLGDD650 pKa = 3.7PTTSGSDD657 pKa = 3.16AFPIPVSNDD666 pKa = 2.88LFNDD670 pKa = 3.48QQQLGGIFGLGMTGLMNNGDD690 pKa = 4.01TNEE693 pKa = 3.88NWINWLDD700 pKa = 3.85RR701 pKa = 11.84RR702 pKa = 11.84DD703 pKa = 4.66DD704 pKa = 3.82PTDD707 pKa = 4.07PNPNDD712 pKa = 3.65VLGGAPGLMTSHH724 pKa = 5.81MTSGTALGVTNTQEE738 pKa = 3.51KK739 pKa = 9.09GYY741 pKa = 10.21QYY743 pKa = 11.03GVQSEE748 pKa = 4.52ASVGSYY754 pKa = 10.55SVVGGMVGFTGGLRR768 pKa = 11.84LYY770 pKa = 10.88GNTAAVGGEE779 pKa = 3.91LGFFIGDD786 pKa = 3.93GTQSNYY792 pKa = 9.98IKK794 pKa = 10.23FTVNVDD800 pKa = 3.34GFEE803 pKa = 4.48MIQEE807 pKa = 4.24INDD810 pKa = 3.38IPQAPITSTLLEE822 pKa = 4.13SDD824 pKa = 3.99RR825 pKa = 11.84PATTGGIRR833 pKa = 11.84FYY835 pKa = 11.23FLIEE839 pKa = 4.2PSTGDD844 pKa = 3.12VTCQYY849 pKa = 10.86SIDD852 pKa = 3.84GGAKK856 pKa = 8.39VTVGTITAQGTILEE870 pKa = 5.4AIQQSTKK877 pKa = 10.89DD878 pKa = 3.72LAVGLIGTSGVSGVEE893 pKa = 5.33LEE895 pKa = 4.49GTWDD899 pKa = 3.55FLNVLFEE906 pKa = 4.4EE907 pKa = 4.72PFVVQSIPDD916 pKa = 3.33IEE918 pKa = 4.57RR919 pKa = 11.84PLGEE923 pKa = 4.13TQEE926 pKa = 4.4ILDD929 pKa = 3.98LSNYY933 pKa = 9.76FDD935 pKa = 6.27DD936 pKa = 4.81NDD938 pKa = 3.72GVANLTYY945 pKa = 10.5SVQNNTNANFTPVITSSQLDD965 pKa = 3.27IALPNLIEE973 pKa = 4.39STDD976 pKa = 3.57LTIRR980 pKa = 11.84AIDD983 pKa = 3.25QGGFFVDD990 pKa = 3.59EE991 pKa = 4.6TFTISINNGPIVLYY1005 pKa = 10.02RR1006 pKa = 11.84VNAGGPEE1013 pKa = 3.79VTAIDD1018 pKa = 4.8GLLNWDD1024 pKa = 4.34EE1025 pKa = 4.68DD1026 pKa = 4.26TDD1028 pKa = 4.44LSPSIYY1034 pKa = 10.55LNEE1037 pKa = 4.13AASNRR1042 pKa = 11.84VFSGSVNALDD1052 pKa = 3.91PSVNVGSVPLGLFTTEE1068 pKa = 4.81RR1069 pKa = 11.84FDD1071 pKa = 5.77DD1072 pKa = 4.04SAGTPNMSYY1081 pKa = 10.77AFPVAMSGNYY1091 pKa = 9.53EE1092 pKa = 3.36IRR1094 pKa = 11.84LYY1096 pKa = 10.32MGEE1099 pKa = 4.24GFIADD1104 pKa = 3.68SQIGQRR1110 pKa = 11.84IFDD1113 pKa = 3.77VQIEE1117 pKa = 5.08GITLPLLNDD1126 pKa = 3.09IDD1128 pKa = 4.15LAEE1131 pKa = 4.55SFGSFNGGVFSHH1143 pKa = 7.26IIKK1146 pKa = 10.73VNDD1149 pKa = 3.34SEE1151 pKa = 4.53INIEE1155 pKa = 4.44FIHH1158 pKa = 6.51GVIEE1162 pKa = 4.2NPMVNAIEE1170 pKa = 4.28ILDD1173 pKa = 3.91VPDD1176 pKa = 4.65FDD1178 pKa = 4.02TPIYY1182 pKa = 10.01LKK1184 pKa = 10.79QIADD1188 pKa = 3.88QINEE1192 pKa = 4.32PGTQINNGLAVQAFGGDD1209 pKa = 3.6GNLSYY1214 pKa = 11.05DD1215 pKa = 4.5ALNLPPGLTIEE1226 pKa = 4.24PTNGLFTGTIALNAADD1242 pKa = 3.95NSPYY1246 pKa = 10.68LVTINVEE1253 pKa = 4.12DD1254 pKa = 3.55SDD1256 pKa = 4.54GFNDD1260 pKa = 4.84DD1261 pKa = 3.41GASIQFLWNINDD1273 pKa = 3.63TSDD1276 pKa = 3.27FRR1278 pKa = 11.84INAGGISFNTPNLGANWEE1296 pKa = 4.34DD1297 pKa = 3.82NATEE1301 pKa = 4.38GAQVGNGFTVNTGRR1315 pKa = 11.84VDD1317 pKa = 4.97PITIDD1322 pKa = 3.49FANRR1326 pKa = 11.84DD1327 pKa = 3.81QASMPAYY1334 pKa = 9.49IDD1336 pKa = 3.09QTAFDD1341 pKa = 3.96MLFAQEE1347 pKa = 4.83RR1348 pKa = 11.84YY1349 pKa = 10.25DD1350 pKa = 3.56IASGDD1355 pKa = 3.62DD1356 pKa = 3.39MAYY1359 pKa = 9.68EE1360 pKa = 4.02IPIANGSFVVNLYY1373 pKa = 9.44FANFFEE1379 pKa = 4.67GTNQIGDD1386 pKa = 3.76RR1387 pKa = 11.84VFDD1390 pKa = 3.72ILLEE1394 pKa = 4.37GAVSRR1399 pKa = 11.84DD1400 pKa = 3.12NFDD1403 pKa = 3.03IVQEE1407 pKa = 4.38FGHH1410 pKa = 6.34LTGGMISLPVDD1421 pKa = 3.61VVDD1424 pKa = 4.46GVLDD1428 pKa = 3.4IGFGRR1433 pKa = 11.84VTQNPLINAIEE1444 pKa = 4.38IYY1446 pKa = 10.37RR1447 pKa = 11.84IDD1449 pKa = 3.65SALDD1453 pKa = 3.3PVVLNNPGNQLNDD1466 pKa = 3.21VNNFISLQINATGGDD1481 pKa = 3.56VMEE1484 pKa = 4.61SFVYY1488 pKa = 9.97FALGLPEE1495 pKa = 4.79GLSIDD1500 pKa = 4.14PNTGEE1505 pKa = 3.71ISGTIAEE1512 pKa = 4.52SASTGGPANDD1522 pKa = 3.91GQHH1525 pKa = 5.75PVKK1528 pKa = 10.4VTVLKK1533 pKa = 10.57EE1534 pKa = 3.91GSAPSSIEE1542 pKa = 4.32FIWEE1546 pKa = 3.89IASSWTEE1553 pKa = 3.34KK1554 pKa = 11.23DD1555 pKa = 2.81EE1556 pKa = 4.42DD1557 pKa = 3.84LNYY1560 pKa = 10.5SARR1563 pKa = 11.84HH1564 pKa = 5.12EE1565 pKa = 4.41CSFVQAGDD1573 pKa = 3.14KK1574 pKa = 10.25FYY1576 pKa = 11.39LMGGRR1581 pKa = 11.84EE1582 pKa = 4.16SATNIDD1588 pKa = 3.43IYY1590 pKa = 11.07DD1591 pKa = 3.73YY1592 pKa = 11.64SSNTWQTLSNSAPFEE1607 pKa = 4.25FNHH1610 pKa = 5.54YY1611 pKa = 9.99QAVTYY1616 pKa = 10.1QGYY1619 pKa = 9.18IWIIGAFKK1627 pKa = 10.86DD1628 pKa = 3.85NNFPTEE1634 pKa = 3.99APAEE1638 pKa = 4.34YY1639 pKa = 9.31IWLFDD1644 pKa = 4.07PVNQNWIQGPEE1655 pKa = 3.63IPTARR1660 pKa = 11.84RR1661 pKa = 11.84RR1662 pKa = 11.84GSAGLVMYY1670 pKa = 10.38NDD1672 pKa = 3.31KK1673 pKa = 10.91FYY1675 pKa = 11.47VVAGNTVGHH1684 pKa = 7.31DD1685 pKa = 3.66GGAVPYY1691 pKa = 10.49LDD1693 pKa = 5.06EE1694 pKa = 4.9YY1695 pKa = 11.48DD1696 pKa = 4.21PKK1698 pKa = 10.55TGTWTVLADD1707 pKa = 3.4APRR1710 pKa = 11.84ARR1712 pKa = 11.84DD1713 pKa = 3.28HH1714 pKa = 6.81FGAVLIDD1721 pKa = 3.68DD1722 pKa = 5.44KK1723 pKa = 11.34IYY1725 pKa = 10.02VASGRR1730 pKa = 11.84TSGGTGGVFAPVIPEE1745 pKa = 3.6VDD1747 pKa = 3.43VYY1749 pKa = 11.49NITNATWSTLPAEE1762 pKa = 5.07DD1763 pKa = 5.32NIPTPRR1769 pKa = 11.84AGTAVANFNNRR1780 pKa = 11.84LVVIGGEE1787 pKa = 4.31VKK1789 pKa = 10.61NEE1791 pKa = 3.65LVYY1794 pKa = 10.92GVVVNDD1800 pKa = 3.71ALKK1803 pKa = 9.24ITEE1806 pKa = 4.29QYY1808 pKa = 11.5DD1809 pKa = 3.39PATEE1813 pKa = 3.82SWTRR1817 pKa = 11.84LPDD1820 pKa = 3.45MNFEE1824 pKa = 4.16RR1825 pKa = 11.84HH1826 pKa = 4.33GTQAIVSGDD1835 pKa = 3.92GIYY1838 pKa = 10.94VLAGSPSRR1846 pKa = 11.84GGGNQKK1852 pKa = 10.33NMEE1855 pKa = 4.2FFGTDD1860 pKa = 3.21NANGAALQGSVIEE1873 pKa = 4.48APAGVFVADD1882 pKa = 3.91NSTSSFQISIVNGNTGTWIDD1902 pKa = 3.36RR1903 pKa = 11.84MEE1905 pKa = 4.05ITGVNAADD1913 pKa = 3.78YY1914 pKa = 10.55SISTGEE1920 pKa = 4.06LTSVLLKK1927 pKa = 10.34QDD1929 pKa = 4.03DD1930 pKa = 3.92LHH1932 pKa = 6.74TIEE1935 pKa = 5.1VNLTGTGEE1943 pKa = 3.91GRR1945 pKa = 11.84NAVLTIYY1952 pKa = 10.73YY1953 pKa = 10.1GLNDD1957 pKa = 3.49SVAINLSNNDD1967 pKa = 3.13VPPIVTNPGNQTNTEE1982 pKa = 4.17GDD1984 pKa = 4.1SVSLQIDD1991 pKa = 3.38ATDD1994 pKa = 3.58NGVSLTYY2001 pKa = 10.21EE2002 pKa = 4.15ATNLPPTLTIDD2013 pKa = 3.76EE2014 pKa = 4.34NSGLITGTIEE2024 pKa = 4.5VANQGDD2030 pKa = 4.35KK2031 pKa = 10.71IFQEE2035 pKa = 4.26EE2036 pKa = 3.95NGYY2039 pKa = 10.13LLIEE2043 pKa = 4.5AEE2045 pKa = 4.36SANLVGGWSLTTVGGAVGITPDD2067 pKa = 3.63SQHH2070 pKa = 6.46FGNPLGGGQLTYY2082 pKa = 10.54KK2083 pKa = 10.51INITNTGVYY2092 pKa = 9.6RR2093 pKa = 11.84FNWRR2097 pKa = 11.84NLYY2100 pKa = 9.16TGNLAAEE2107 pKa = 4.88NNDD2110 pKa = 2.86SWLKK2114 pKa = 10.47FPNDD2118 pKa = 3.46SNVVFFGYY2126 pKa = 10.66DD2127 pKa = 2.83GFTTSNEE2134 pKa = 4.26SIYY2137 pKa = 11.14TNIIEE2142 pKa = 4.2NDD2144 pKa = 3.38IYY2146 pKa = 11.65DD2147 pKa = 4.26DD2148 pKa = 3.59LCYY2151 pKa = 10.36PKK2153 pKa = 10.9GSGLGNNITTDD2164 pKa = 3.19MGGTLPLGLGSNGYY2178 pKa = 9.64FKK2180 pKa = 10.97VYY2182 pKa = 9.21RR2183 pKa = 11.84TGGQPEE2189 pKa = 4.33VYY2191 pKa = 8.64EE2192 pKa = 4.27WEE2194 pKa = 4.59SKK2196 pKa = 11.04VSDD2199 pKa = 4.08GDD2201 pKa = 3.7DD2202 pKa = 3.01HH2203 pKa = 9.39SVFVWFKK2210 pKa = 10.6NPGVYY2215 pKa = 10.14EE2216 pKa = 4.58FIVSEE2221 pKa = 4.21RR2222 pKa = 11.84SSGHH2226 pKa = 6.68IIDD2229 pKa = 5.08KK2230 pKa = 9.74MVLYY2234 pKa = 10.49KK2235 pKa = 9.62VTQAGDD2241 pKa = 3.46FNTEE2245 pKa = 3.76AKK2247 pKa = 10.44LDD2249 pKa = 4.07ALPEE2253 pKa = 4.69SNFTGSSGGAADD2265 pKa = 3.55NSPYY2269 pKa = 10.49NVQVTVTDD2277 pKa = 4.7DD2278 pKa = 3.59SDD2280 pKa = 5.06LSDD2283 pKa = 3.53TVDD2286 pKa = 4.03FTWFVGSLGQSLFAVAEE2303 pKa = 4.12SDD2305 pKa = 3.07ITTGNSPLSVNFVGSNSFDD2324 pKa = 4.88DD2325 pKa = 3.84IAVTGYY2331 pKa = 10.8LWDD2334 pKa = 4.28FQDD2337 pKa = 4.45GNTSTEE2343 pKa = 3.92ANPTYY2348 pKa = 10.9VFNSSGTYY2356 pKa = 8.96EE2357 pKa = 4.02VSLTVIDD2364 pKa = 4.41GDD2366 pKa = 4.38GNTDD2370 pKa = 3.14TDD2372 pKa = 3.36ILTIEE2377 pKa = 4.36VQEE2380 pKa = 4.25PLPPTSVAEE2389 pKa = 4.24TNITAGNAPLEE2400 pKa = 4.17VSFTGSNSFDD2410 pKa = 3.77DD2411 pKa = 4.19KK2412 pKa = 10.95PNNIISYY2419 pKa = 9.69AWDD2422 pKa = 3.9FGTGDD2427 pKa = 3.56TSVEE2431 pKa = 3.76QDD2433 pKa = 3.18PTYY2436 pKa = 10.5TFLSGGVYY2444 pKa = 8.99TVSLTVEE2451 pKa = 4.6DD2452 pKa = 4.58GQGLTDD2458 pKa = 3.55TATLEE2463 pKa = 4.06ITVNGAPVAIATADD2477 pKa = 3.5VTSGNEE2483 pKa = 3.59ALEE2486 pKa = 4.28VNFTGSGSTDD2496 pKa = 3.57DD2497 pKa = 4.35NNDD2500 pKa = 2.94IVSYY2504 pKa = 10.69AWDD2507 pKa = 4.09FGTGDD2512 pKa = 3.9MSTEE2516 pKa = 3.91ANPTYY2521 pKa = 10.46TFTTGGVYY2529 pKa = 9.06TVSLTVV2535 pKa = 3.12
MM1 pKa = 7.13NQQFLNPIARR11 pKa = 11.84SMVQVVKK18 pKa = 10.94KK19 pKa = 10.63NLLLYY24 pKa = 10.8GLLLAFGFYY33 pKa = 9.49QANAQVNFDD42 pKa = 3.51QSTLNFNGIGEE53 pKa = 4.47STNGFTSIMFGPDD66 pKa = 2.38GRR68 pKa = 11.84LYY70 pKa = 9.2VAEE73 pKa = 3.86YY74 pKa = 10.11HH75 pKa = 6.23GLIKK79 pKa = 10.41IFTIEE84 pKa = 4.17RR85 pKa = 11.84QSSTSYY91 pKa = 9.94TVTAVEE97 pKa = 4.5EE98 pKa = 4.23LNGIQTMEE106 pKa = 4.31DD107 pKa = 3.6HH108 pKa = 7.05NDD110 pKa = 3.61DD111 pKa = 3.7GSGFSSLEE119 pKa = 3.91RR120 pKa = 11.84EE121 pKa = 4.47TTGLTVAGTATNPIIYY137 pKa = 8.21VTSSDD142 pKa = 3.51FRR144 pKa = 11.84IGSGPAGGNGDD155 pKa = 3.66VGLDD159 pKa = 3.63TNSGVITRR167 pKa = 11.84FTWTGSAWDD176 pKa = 3.55VVDD179 pKa = 5.28LVRR182 pKa = 11.84GLPRR186 pKa = 11.84SEE188 pKa = 4.39EE189 pKa = 3.77NHH191 pKa = 5.43ATNGLEE197 pKa = 3.86YY198 pKa = 10.89AQINGVNYY206 pKa = 10.59LLVTSGGITNAGSPSTNFVFTCEE229 pKa = 4.04YY230 pKa = 9.98ALSGAILSVNLDD242 pKa = 4.42EE243 pKa = 5.82INSLPILTDD252 pKa = 2.84SGRR255 pKa = 11.84QYY257 pKa = 10.9IYY259 pKa = 10.74DD260 pKa = 4.93LPTLDD265 pKa = 5.62DD266 pKa = 3.64PTRR269 pKa = 11.84PNSNGIIDD277 pKa = 4.57PNDD280 pKa = 3.68PGYY283 pKa = 9.57TGEE286 pKa = 5.3DD287 pKa = 3.83LNDD290 pKa = 3.26PWGGNDD296 pKa = 4.14GLNMAIIDD304 pKa = 3.85PAGPVQVFSPGFRR317 pKa = 11.84NAYY320 pKa = 9.45DD321 pKa = 3.63LVVTEE326 pKa = 4.77SGAVYY331 pKa = 8.72ATDD334 pKa = 3.32NGANGGWGGFPVNEE348 pKa = 4.16GTVNANNDD356 pKa = 3.48YY357 pKa = 11.2DD358 pKa = 4.18PNEE361 pKa = 4.4TGSTFATQAPDD372 pKa = 3.36GEE374 pKa = 4.86RR375 pKa = 11.84ILNQDD380 pKa = 3.52HH381 pKa = 7.1LEE383 pKa = 4.68LITTDD388 pKa = 3.42LSSYY392 pKa = 10.47TFGSYY397 pKa = 9.89YY398 pKa = 10.44AGHH401 pKa = 7.38PNPVRR406 pKa = 11.84ANPSGAGLYY415 pKa = 8.17TVNDD419 pKa = 4.13TNSVWRR425 pKa = 11.84TLIYY429 pKa = 10.86DD430 pKa = 4.09PDD432 pKa = 4.16GSSPNSTTDD441 pKa = 3.01VTLGLPANWPPVNTANPDD459 pKa = 3.31EE460 pKa = 4.74GDD462 pKa = 3.01WRR464 pKa = 11.84GPTTPNPDD472 pKa = 3.43GPDD475 pKa = 3.46NSPVLVWGTNTNGIDD490 pKa = 3.69EE491 pKa = 4.38YY492 pKa = 10.69TATNFAGQMQGNLIAGVNSGVLRR515 pKa = 11.84RR516 pKa = 11.84VEE518 pKa = 4.3LNPDD522 pKa = 2.92GSVANFTAEE531 pKa = 4.11FFSGLGGNALAITCNSDD548 pKa = 3.29TEE550 pKa = 4.42IFPGTIWAGTLGGDD564 pKa = 4.53LIVFEE569 pKa = 4.6PQDD572 pKa = 3.81FVVCILPGEE581 pKa = 4.17MDD583 pKa = 4.32YY584 pKa = 11.52DD585 pKa = 4.0PSADD589 pKa = 3.86NDD591 pKa = 3.81SDD593 pKa = 5.24GYY595 pKa = 9.96TNDD598 pKa = 5.87DD599 pKa = 3.58EE600 pKa = 5.03DD601 pKa = 4.75QNGTDD606 pKa = 3.61LCNGGSQPSDD616 pKa = 3.14FDD618 pKa = 3.68KK619 pKa = 11.25AAGGILVSDD628 pKa = 5.05LLDD631 pKa = 4.07NDD633 pKa = 4.93DD634 pKa = 5.07DD635 pKa = 4.94FDD637 pKa = 6.86GIVDD641 pKa = 3.92SADD644 pKa = 3.37PFQLGDD650 pKa = 3.7PTTSGSDD657 pKa = 3.16AFPIPVSNDD666 pKa = 2.88LFNDD670 pKa = 3.48QQQLGGIFGLGMTGLMNNGDD690 pKa = 4.01TNEE693 pKa = 3.88NWINWLDD700 pKa = 3.85RR701 pKa = 11.84RR702 pKa = 11.84DD703 pKa = 4.66DD704 pKa = 3.82PTDD707 pKa = 4.07PNPNDD712 pKa = 3.65VLGGAPGLMTSHH724 pKa = 5.81MTSGTALGVTNTQEE738 pKa = 3.51KK739 pKa = 9.09GYY741 pKa = 10.21QYY743 pKa = 11.03GVQSEE748 pKa = 4.52ASVGSYY754 pKa = 10.55SVVGGMVGFTGGLRR768 pKa = 11.84LYY770 pKa = 10.88GNTAAVGGEE779 pKa = 3.91LGFFIGDD786 pKa = 3.93GTQSNYY792 pKa = 9.98IKK794 pKa = 10.23FTVNVDD800 pKa = 3.34GFEE803 pKa = 4.48MIQEE807 pKa = 4.24INDD810 pKa = 3.38IPQAPITSTLLEE822 pKa = 4.13SDD824 pKa = 3.99RR825 pKa = 11.84PATTGGIRR833 pKa = 11.84FYY835 pKa = 11.23FLIEE839 pKa = 4.2PSTGDD844 pKa = 3.12VTCQYY849 pKa = 10.86SIDD852 pKa = 3.84GGAKK856 pKa = 8.39VTVGTITAQGTILEE870 pKa = 5.4AIQQSTKK877 pKa = 10.89DD878 pKa = 3.72LAVGLIGTSGVSGVEE893 pKa = 5.33LEE895 pKa = 4.49GTWDD899 pKa = 3.55FLNVLFEE906 pKa = 4.4EE907 pKa = 4.72PFVVQSIPDD916 pKa = 3.33IEE918 pKa = 4.57RR919 pKa = 11.84PLGEE923 pKa = 4.13TQEE926 pKa = 4.4ILDD929 pKa = 3.98LSNYY933 pKa = 9.76FDD935 pKa = 6.27DD936 pKa = 4.81NDD938 pKa = 3.72GVANLTYY945 pKa = 10.5SVQNNTNANFTPVITSSQLDD965 pKa = 3.27IALPNLIEE973 pKa = 4.39STDD976 pKa = 3.57LTIRR980 pKa = 11.84AIDD983 pKa = 3.25QGGFFVDD990 pKa = 3.59EE991 pKa = 4.6TFTISINNGPIVLYY1005 pKa = 10.02RR1006 pKa = 11.84VNAGGPEE1013 pKa = 3.79VTAIDD1018 pKa = 4.8GLLNWDD1024 pKa = 4.34EE1025 pKa = 4.68DD1026 pKa = 4.26TDD1028 pKa = 4.44LSPSIYY1034 pKa = 10.55LNEE1037 pKa = 4.13AASNRR1042 pKa = 11.84VFSGSVNALDD1052 pKa = 3.91PSVNVGSVPLGLFTTEE1068 pKa = 4.81RR1069 pKa = 11.84FDD1071 pKa = 5.77DD1072 pKa = 4.04SAGTPNMSYY1081 pKa = 10.77AFPVAMSGNYY1091 pKa = 9.53EE1092 pKa = 3.36IRR1094 pKa = 11.84LYY1096 pKa = 10.32MGEE1099 pKa = 4.24GFIADD1104 pKa = 3.68SQIGQRR1110 pKa = 11.84IFDD1113 pKa = 3.77VQIEE1117 pKa = 5.08GITLPLLNDD1126 pKa = 3.09IDD1128 pKa = 4.15LAEE1131 pKa = 4.55SFGSFNGGVFSHH1143 pKa = 7.26IIKK1146 pKa = 10.73VNDD1149 pKa = 3.34SEE1151 pKa = 4.53INIEE1155 pKa = 4.44FIHH1158 pKa = 6.51GVIEE1162 pKa = 4.2NPMVNAIEE1170 pKa = 4.28ILDD1173 pKa = 3.91VPDD1176 pKa = 4.65FDD1178 pKa = 4.02TPIYY1182 pKa = 10.01LKK1184 pKa = 10.79QIADD1188 pKa = 3.88QINEE1192 pKa = 4.32PGTQINNGLAVQAFGGDD1209 pKa = 3.6GNLSYY1214 pKa = 11.05DD1215 pKa = 4.5ALNLPPGLTIEE1226 pKa = 4.24PTNGLFTGTIALNAADD1242 pKa = 3.95NSPYY1246 pKa = 10.68LVTINVEE1253 pKa = 4.12DD1254 pKa = 3.55SDD1256 pKa = 4.54GFNDD1260 pKa = 4.84DD1261 pKa = 3.41GASIQFLWNINDD1273 pKa = 3.63TSDD1276 pKa = 3.27FRR1278 pKa = 11.84INAGGISFNTPNLGANWEE1296 pKa = 4.34DD1297 pKa = 3.82NATEE1301 pKa = 4.38GAQVGNGFTVNTGRR1315 pKa = 11.84VDD1317 pKa = 4.97PITIDD1322 pKa = 3.49FANRR1326 pKa = 11.84DD1327 pKa = 3.81QASMPAYY1334 pKa = 9.49IDD1336 pKa = 3.09QTAFDD1341 pKa = 3.96MLFAQEE1347 pKa = 4.83RR1348 pKa = 11.84YY1349 pKa = 10.25DD1350 pKa = 3.56IASGDD1355 pKa = 3.62DD1356 pKa = 3.39MAYY1359 pKa = 9.68EE1360 pKa = 4.02IPIANGSFVVNLYY1373 pKa = 9.44FANFFEE1379 pKa = 4.67GTNQIGDD1386 pKa = 3.76RR1387 pKa = 11.84VFDD1390 pKa = 3.72ILLEE1394 pKa = 4.37GAVSRR1399 pKa = 11.84DD1400 pKa = 3.12NFDD1403 pKa = 3.03IVQEE1407 pKa = 4.38FGHH1410 pKa = 6.34LTGGMISLPVDD1421 pKa = 3.61VVDD1424 pKa = 4.46GVLDD1428 pKa = 3.4IGFGRR1433 pKa = 11.84VTQNPLINAIEE1444 pKa = 4.38IYY1446 pKa = 10.37RR1447 pKa = 11.84IDD1449 pKa = 3.65SALDD1453 pKa = 3.3PVVLNNPGNQLNDD1466 pKa = 3.21VNNFISLQINATGGDD1481 pKa = 3.56VMEE1484 pKa = 4.61SFVYY1488 pKa = 9.97FALGLPEE1495 pKa = 4.79GLSIDD1500 pKa = 4.14PNTGEE1505 pKa = 3.71ISGTIAEE1512 pKa = 4.52SASTGGPANDD1522 pKa = 3.91GQHH1525 pKa = 5.75PVKK1528 pKa = 10.4VTVLKK1533 pKa = 10.57EE1534 pKa = 3.91GSAPSSIEE1542 pKa = 4.32FIWEE1546 pKa = 3.89IASSWTEE1553 pKa = 3.34KK1554 pKa = 11.23DD1555 pKa = 2.81EE1556 pKa = 4.42DD1557 pKa = 3.84LNYY1560 pKa = 10.5SARR1563 pKa = 11.84HH1564 pKa = 5.12EE1565 pKa = 4.41CSFVQAGDD1573 pKa = 3.14KK1574 pKa = 10.25FYY1576 pKa = 11.39LMGGRR1581 pKa = 11.84EE1582 pKa = 4.16SATNIDD1588 pKa = 3.43IYY1590 pKa = 11.07DD1591 pKa = 3.73YY1592 pKa = 11.64SSNTWQTLSNSAPFEE1607 pKa = 4.25FNHH1610 pKa = 5.54YY1611 pKa = 9.99QAVTYY1616 pKa = 10.1QGYY1619 pKa = 9.18IWIIGAFKK1627 pKa = 10.86DD1628 pKa = 3.85NNFPTEE1634 pKa = 3.99APAEE1638 pKa = 4.34YY1639 pKa = 9.31IWLFDD1644 pKa = 4.07PVNQNWIQGPEE1655 pKa = 3.63IPTARR1660 pKa = 11.84RR1661 pKa = 11.84RR1662 pKa = 11.84GSAGLVMYY1670 pKa = 10.38NDD1672 pKa = 3.31KK1673 pKa = 10.91FYY1675 pKa = 11.47VVAGNTVGHH1684 pKa = 7.31DD1685 pKa = 3.66GGAVPYY1691 pKa = 10.49LDD1693 pKa = 5.06EE1694 pKa = 4.9YY1695 pKa = 11.48DD1696 pKa = 4.21PKK1698 pKa = 10.55TGTWTVLADD1707 pKa = 3.4APRR1710 pKa = 11.84ARR1712 pKa = 11.84DD1713 pKa = 3.28HH1714 pKa = 6.81FGAVLIDD1721 pKa = 3.68DD1722 pKa = 5.44KK1723 pKa = 11.34IYY1725 pKa = 10.02VASGRR1730 pKa = 11.84TSGGTGGVFAPVIPEE1745 pKa = 3.6VDD1747 pKa = 3.43VYY1749 pKa = 11.49NITNATWSTLPAEE1762 pKa = 5.07DD1763 pKa = 5.32NIPTPRR1769 pKa = 11.84AGTAVANFNNRR1780 pKa = 11.84LVVIGGEE1787 pKa = 4.31VKK1789 pKa = 10.61NEE1791 pKa = 3.65LVYY1794 pKa = 10.92GVVVNDD1800 pKa = 3.71ALKK1803 pKa = 9.24ITEE1806 pKa = 4.29QYY1808 pKa = 11.5DD1809 pKa = 3.39PATEE1813 pKa = 3.82SWTRR1817 pKa = 11.84LPDD1820 pKa = 3.45MNFEE1824 pKa = 4.16RR1825 pKa = 11.84HH1826 pKa = 4.33GTQAIVSGDD1835 pKa = 3.92GIYY1838 pKa = 10.94VLAGSPSRR1846 pKa = 11.84GGGNQKK1852 pKa = 10.33NMEE1855 pKa = 4.2FFGTDD1860 pKa = 3.21NANGAALQGSVIEE1873 pKa = 4.48APAGVFVADD1882 pKa = 3.91NSTSSFQISIVNGNTGTWIDD1902 pKa = 3.36RR1903 pKa = 11.84MEE1905 pKa = 4.05ITGVNAADD1913 pKa = 3.78YY1914 pKa = 10.55SISTGEE1920 pKa = 4.06LTSVLLKK1927 pKa = 10.34QDD1929 pKa = 4.03DD1930 pKa = 3.92LHH1932 pKa = 6.74TIEE1935 pKa = 5.1VNLTGTGEE1943 pKa = 3.91GRR1945 pKa = 11.84NAVLTIYY1952 pKa = 10.73YY1953 pKa = 10.1GLNDD1957 pKa = 3.49SVAINLSNNDD1967 pKa = 3.13VPPIVTNPGNQTNTEE1982 pKa = 4.17GDD1984 pKa = 4.1SVSLQIDD1991 pKa = 3.38ATDD1994 pKa = 3.58NGVSLTYY2001 pKa = 10.21EE2002 pKa = 4.15ATNLPPTLTIDD2013 pKa = 3.76EE2014 pKa = 4.34NSGLITGTIEE2024 pKa = 4.5VANQGDD2030 pKa = 4.35KK2031 pKa = 10.71IFQEE2035 pKa = 4.26EE2036 pKa = 3.95NGYY2039 pKa = 10.13LLIEE2043 pKa = 4.5AEE2045 pKa = 4.36SANLVGGWSLTTVGGAVGITPDD2067 pKa = 3.63SQHH2070 pKa = 6.46FGNPLGGGQLTYY2082 pKa = 10.54KK2083 pKa = 10.51INITNTGVYY2092 pKa = 9.6RR2093 pKa = 11.84FNWRR2097 pKa = 11.84NLYY2100 pKa = 9.16TGNLAAEE2107 pKa = 4.88NNDD2110 pKa = 2.86SWLKK2114 pKa = 10.47FPNDD2118 pKa = 3.46SNVVFFGYY2126 pKa = 10.66DD2127 pKa = 2.83GFTTSNEE2134 pKa = 4.26SIYY2137 pKa = 11.14TNIIEE2142 pKa = 4.2NDD2144 pKa = 3.38IYY2146 pKa = 11.65DD2147 pKa = 4.26DD2148 pKa = 3.59LCYY2151 pKa = 10.36PKK2153 pKa = 10.9GSGLGNNITTDD2164 pKa = 3.19MGGTLPLGLGSNGYY2178 pKa = 9.64FKK2180 pKa = 10.97VYY2182 pKa = 9.21RR2183 pKa = 11.84TGGQPEE2189 pKa = 4.33VYY2191 pKa = 8.64EE2192 pKa = 4.27WEE2194 pKa = 4.59SKK2196 pKa = 11.04VSDD2199 pKa = 4.08GDD2201 pKa = 3.7DD2202 pKa = 3.01HH2203 pKa = 9.39SVFVWFKK2210 pKa = 10.6NPGVYY2215 pKa = 10.14EE2216 pKa = 4.58FIVSEE2221 pKa = 4.21RR2222 pKa = 11.84SSGHH2226 pKa = 6.68IIDD2229 pKa = 5.08KK2230 pKa = 9.74MVLYY2234 pKa = 10.49KK2235 pKa = 9.62VTQAGDD2241 pKa = 3.46FNTEE2245 pKa = 3.76AKK2247 pKa = 10.44LDD2249 pKa = 4.07ALPEE2253 pKa = 4.69SNFTGSSGGAADD2265 pKa = 3.55NSPYY2269 pKa = 10.49NVQVTVTDD2277 pKa = 4.7DD2278 pKa = 3.59SDD2280 pKa = 5.06LSDD2283 pKa = 3.53TVDD2286 pKa = 4.03FTWFVGSLGQSLFAVAEE2303 pKa = 4.12SDD2305 pKa = 3.07ITTGNSPLSVNFVGSNSFDD2324 pKa = 4.88DD2325 pKa = 3.84IAVTGYY2331 pKa = 10.8LWDD2334 pKa = 4.28FQDD2337 pKa = 4.45GNTSTEE2343 pKa = 3.92ANPTYY2348 pKa = 10.9VFNSSGTYY2356 pKa = 8.96EE2357 pKa = 4.02VSLTVIDD2364 pKa = 4.41GDD2366 pKa = 4.38GNTDD2370 pKa = 3.14TDD2372 pKa = 3.36ILTIEE2377 pKa = 4.36VQEE2380 pKa = 4.25PLPPTSVAEE2389 pKa = 4.24TNITAGNAPLEE2400 pKa = 4.17VSFTGSNSFDD2410 pKa = 3.77DD2411 pKa = 4.19KK2412 pKa = 10.95PNNIISYY2419 pKa = 9.69AWDD2422 pKa = 3.9FGTGDD2427 pKa = 3.56TSVEE2431 pKa = 3.76QDD2433 pKa = 3.18PTYY2436 pKa = 10.5TFLSGGVYY2444 pKa = 8.99TVSLTVEE2451 pKa = 4.6DD2452 pKa = 4.58GQGLTDD2458 pKa = 3.55TATLEE2463 pKa = 4.06ITVNGAPVAIATADD2477 pKa = 3.5VTSGNEE2483 pKa = 3.59ALEE2486 pKa = 4.28VNFTGSGSTDD2496 pKa = 3.57DD2497 pKa = 4.35NNDD2500 pKa = 2.94IVSYY2504 pKa = 10.69AWDD2507 pKa = 4.09FGTGDD2512 pKa = 3.9MSTEE2516 pKa = 3.91ANPTYY2521 pKa = 10.46TFTTGGVYY2529 pKa = 9.06TVSLTVV2535 pKa = 3.12
Molecular weight: 270.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S7T853|A0A2S7T853_9FLAO Glutamyl-tRNA amidotransferase OS=Aureicoccus marinus OX=754435 GN=BST99_08050 PE=4 SV=1
MM1 pKa = 6.85NQRR4 pKa = 11.84RR5 pKa = 11.84CEE7 pKa = 4.09EE8 pKa = 4.3CNSPLKK14 pKa = 10.68GRR16 pKa = 11.84SDD18 pKa = 3.24KK19 pKa = 10.91RR20 pKa = 11.84FCGDD24 pKa = 3.15YY25 pKa = 10.91CRR27 pKa = 11.84NTYY30 pKa = 10.45HH31 pKa = 6.42NRR33 pKa = 11.84KK34 pKa = 7.0HH35 pKa = 5.24QKK37 pKa = 9.39RR38 pKa = 11.84NRR40 pKa = 11.84VRR42 pKa = 11.84YY43 pKa = 7.67ATHH46 pKa = 6.5QKK48 pKa = 9.95LQRR51 pKa = 11.84NRR53 pKa = 11.84QILKK57 pKa = 10.21EE58 pKa = 3.9ILGSKK63 pKa = 9.17ARR65 pKa = 11.84RR66 pKa = 11.84VIGRR70 pKa = 11.84EE71 pKa = 3.54QLIQLGFDD79 pKa = 3.53FGFLTEE85 pKa = 4.16ILRR88 pKa = 11.84LTRR91 pKa = 11.84GQNYY95 pKa = 9.58YY96 pKa = 10.5YY97 pKa = 9.8IYY99 pKa = 10.64DD100 pKa = 3.23VGYY103 pKa = 10.79RR104 pKa = 11.84LLEE107 pKa = 4.14ANRR110 pKa = 11.84LLIVHH115 pKa = 6.81KK116 pKa = 10.17DD117 pKa = 3.37KK118 pKa = 11.3RR119 pKa = 11.84PLAISSS125 pKa = 3.57
MM1 pKa = 6.85NQRR4 pKa = 11.84RR5 pKa = 11.84CEE7 pKa = 4.09EE8 pKa = 4.3CNSPLKK14 pKa = 10.68GRR16 pKa = 11.84SDD18 pKa = 3.24KK19 pKa = 10.91RR20 pKa = 11.84FCGDD24 pKa = 3.15YY25 pKa = 10.91CRR27 pKa = 11.84NTYY30 pKa = 10.45HH31 pKa = 6.42NRR33 pKa = 11.84KK34 pKa = 7.0HH35 pKa = 5.24QKK37 pKa = 9.39RR38 pKa = 11.84NRR40 pKa = 11.84VRR42 pKa = 11.84YY43 pKa = 7.67ATHH46 pKa = 6.5QKK48 pKa = 9.95LQRR51 pKa = 11.84NRR53 pKa = 11.84QILKK57 pKa = 10.21EE58 pKa = 3.9ILGSKK63 pKa = 9.17ARR65 pKa = 11.84RR66 pKa = 11.84VIGRR70 pKa = 11.84EE71 pKa = 3.54QLIQLGFDD79 pKa = 3.53FGFLTEE85 pKa = 4.16ILRR88 pKa = 11.84LTRR91 pKa = 11.84GQNYY95 pKa = 9.58YY96 pKa = 10.5YY97 pKa = 9.8IYY99 pKa = 10.64DD100 pKa = 3.23VGYY103 pKa = 10.79RR104 pKa = 11.84LLEE107 pKa = 4.14ANRR110 pKa = 11.84LLIVHH115 pKa = 6.81KK116 pKa = 10.17DD117 pKa = 3.37KK118 pKa = 11.3RR119 pKa = 11.84PLAISSS125 pKa = 3.57
Molecular weight: 15.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
833196 |
27 |
4108 |
327.6 |
36.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.087 ± 0.045 | 0.746 ± 0.018 |
5.49 ± 0.045 | 7.141 ± 0.058 |
4.941 ± 0.041 | 7.121 ± 0.05 |
1.863 ± 0.026 | 6.347 ± 0.045 |
6.118 ± 0.062 | 10.222 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.263 ± 0.028 | 4.736 ± 0.046 |
3.92 ± 0.028 | 4.26 ± 0.033 |
4.558 ± 0.042 | 6.491 ± 0.038 |
5.248 ± 0.065 | 6.304 ± 0.041 |
1.265 ± 0.022 | 3.877 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
