
Hubei narna-like virus 22
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.66
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
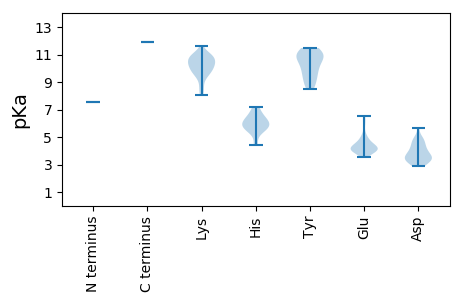
Protein with the lowest isoelectric point:
>tr|A0A1L3KIM3|A0A1L3KIM3_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 22 OX=1922953 PE=4 SV=1
MM1 pKa = 7.58EE2 pKa = 6.53KK3 pKa = 10.29DD4 pKa = 5.69LKK6 pKa = 10.76MKK8 pKa = 8.65WQQRR12 pKa = 11.84EE13 pKa = 3.99WFNPSHH19 pKa = 6.25LTGSSIPGSGKK30 pKa = 10.49AMSIRR35 pKa = 11.84NRR37 pKa = 11.84NGNISYY43 pKa = 10.95ASIVSAYY50 pKa = 10.42SFSLSTAPLFIWKK63 pKa = 9.46FLDD66 pKa = 4.36DD67 pKa = 3.49IKK69 pKa = 10.87YY70 pKa = 9.71PEE72 pKa = 4.25MVYY75 pKa = 10.18EE76 pKa = 4.07VQGNLEE82 pKa = 3.94QSIKK86 pKa = 10.3LSQIIEE92 pKa = 3.9EE93 pKa = 4.09RR94 pKa = 11.84LQNEE98 pKa = 4.15RR99 pKa = 11.84LLDD102 pKa = 3.84EE103 pKa = 4.98ALSGNNFLDD112 pKa = 4.36DD113 pKa = 4.69DD114 pKa = 4.92SFVDD118 pKa = 3.58YY119 pKa = 11.15QKK121 pKa = 10.93FVTPILGLHH130 pKa = 6.28HH131 pKa = 6.75SVGNIGFLQQGSGKK145 pKa = 9.72LRR147 pKa = 11.84AVANPNRR154 pKa = 11.84FVQYY158 pKa = 10.26CNSPLGEE165 pKa = 4.24VLSSHH170 pKa = 5.58QQGLDD175 pKa = 2.93GCFVYY180 pKa = 10.64NQEE183 pKa = 5.88AGIEE187 pKa = 4.13WVKK190 pKa = 11.01RR191 pKa = 11.84KK192 pKa = 9.74LKK194 pKa = 10.35QGNTLASFDD203 pKa = 3.75MSAATDD209 pKa = 3.29RR210 pKa = 11.84LDD212 pKa = 3.33FQKK215 pKa = 10.84FINEE219 pKa = 3.95YY220 pKa = 9.53FVNISDD226 pKa = 3.74EE227 pKa = 4.17RR228 pKa = 11.84HH229 pKa = 5.83PLLSRR234 pKa = 11.84SIEE237 pKa = 4.13LFKK240 pKa = 10.79DD241 pKa = 3.51TSLSPWSLPGFVADD255 pKa = 4.02MLQMPSNEE263 pKa = 4.03ISWKK267 pKa = 9.82IGQPLGLRR275 pKa = 11.84PSFPLLTIMNCAMASQAVRR294 pKa = 11.84NLDD297 pKa = 3.32GYY299 pKa = 9.15GTYY302 pKa = 8.93NTFACVGDD310 pKa = 4.05DD311 pKa = 4.14LVIQAEE317 pKa = 4.23YY318 pKa = 11.0ADD320 pKa = 4.99AYY322 pKa = 8.82VQVVSEE328 pKa = 4.37FSGLINRR335 pKa = 11.84DD336 pKa = 2.98KK337 pKa = 11.6GMTSSKK343 pKa = 10.32YY344 pKa = 10.79AEE346 pKa = 4.51FCSQLITADD355 pKa = 2.9SSYY358 pKa = 11.38ALKK361 pKa = 10.37PRR363 pKa = 11.84WIDD366 pKa = 3.61SIEE369 pKa = 4.02GSLNNIEE376 pKa = 4.5KK377 pKa = 10.11FQTSGLHH384 pKa = 5.53PKK386 pKa = 9.52VPEE389 pKa = 4.56WIQRR393 pKa = 11.84LYY395 pKa = 10.92NAIAKK400 pKa = 10.04RR401 pKa = 11.84SIKK404 pKa = 10.64GFTKK408 pKa = 10.53YY409 pKa = 10.79GYY411 pKa = 8.5TKK413 pKa = 9.78TDD415 pKa = 3.73CYY417 pKa = 11.48DD418 pKa = 3.42SLKK421 pKa = 11.04DD422 pKa = 3.36RR423 pKa = 11.84VAIHH427 pKa = 7.15DD428 pKa = 4.85FIQSTRR434 pKa = 11.84KK435 pKa = 9.38APRR438 pKa = 11.84DD439 pKa = 3.33IEE441 pKa = 4.79EE442 pKa = 4.01ITLQTLYY449 pKa = 9.94MRR451 pKa = 11.84AVSEE455 pKa = 4.32QDD457 pKa = 3.2EE458 pKa = 4.63LNQKK462 pKa = 8.05VKK464 pKa = 10.84CFDD467 pKa = 4.8LSTSWAMPKK476 pKa = 10.36LGQNKK481 pKa = 8.6QEE483 pKa = 3.54ISTRR487 pKa = 11.84HH488 pKa = 5.62KK489 pKa = 11.03DD490 pKa = 2.94IVQKK494 pKa = 11.06YY495 pKa = 9.57DD496 pKa = 3.16IEE498 pKa = 6.22LGDD501 pKa = 4.35LPSDD505 pKa = 3.52RR506 pKa = 11.84ATSVAVPGIQQWNYY520 pKa = 10.97KK521 pKa = 8.18EE522 pKa = 5.08DD523 pKa = 3.92CYY525 pKa = 11.3SPKK528 pKa = 10.19VSEE531 pKa = 4.71LNTAKK536 pKa = 10.84KK537 pKa = 10.02LLKK540 pKa = 9.46TIEE543 pKa = 4.33SIKK546 pKa = 10.57VEE548 pKa = 4.15SAGTLISASQEE559 pKa = 4.03TKK561 pKa = 9.89HH562 pKa = 6.31CRR564 pKa = 11.84TEE566 pKa = 3.64ILTDD570 pKa = 4.06LSNHH574 pKa = 5.96EE575 pKa = 4.36SVIVHH580 pKa = 5.85TNKK583 pKa = 9.09ATNTTAIQTKK593 pKa = 9.81HH594 pKa = 4.41NQKK597 pKa = 10.34SKK599 pKa = 10.76RR600 pKa = 11.84LEE602 pKa = 4.12DD603 pKa = 3.54LLANLKK609 pKa = 9.93IDD611 pKa = 4.47DD612 pKa = 4.63PTDD615 pKa = 3.35EE616 pKa = 5.11AQTSMEE622 pKa = 3.87TSYY625 pKa = 11.91
MM1 pKa = 7.58EE2 pKa = 6.53KK3 pKa = 10.29DD4 pKa = 5.69LKK6 pKa = 10.76MKK8 pKa = 8.65WQQRR12 pKa = 11.84EE13 pKa = 3.99WFNPSHH19 pKa = 6.25LTGSSIPGSGKK30 pKa = 10.49AMSIRR35 pKa = 11.84NRR37 pKa = 11.84NGNISYY43 pKa = 10.95ASIVSAYY50 pKa = 10.42SFSLSTAPLFIWKK63 pKa = 9.46FLDD66 pKa = 4.36DD67 pKa = 3.49IKK69 pKa = 10.87YY70 pKa = 9.71PEE72 pKa = 4.25MVYY75 pKa = 10.18EE76 pKa = 4.07VQGNLEE82 pKa = 3.94QSIKK86 pKa = 10.3LSQIIEE92 pKa = 3.9EE93 pKa = 4.09RR94 pKa = 11.84LQNEE98 pKa = 4.15RR99 pKa = 11.84LLDD102 pKa = 3.84EE103 pKa = 4.98ALSGNNFLDD112 pKa = 4.36DD113 pKa = 4.69DD114 pKa = 4.92SFVDD118 pKa = 3.58YY119 pKa = 11.15QKK121 pKa = 10.93FVTPILGLHH130 pKa = 6.28HH131 pKa = 6.75SVGNIGFLQQGSGKK145 pKa = 9.72LRR147 pKa = 11.84AVANPNRR154 pKa = 11.84FVQYY158 pKa = 10.26CNSPLGEE165 pKa = 4.24VLSSHH170 pKa = 5.58QQGLDD175 pKa = 2.93GCFVYY180 pKa = 10.64NQEE183 pKa = 5.88AGIEE187 pKa = 4.13WVKK190 pKa = 11.01RR191 pKa = 11.84KK192 pKa = 9.74LKK194 pKa = 10.35QGNTLASFDD203 pKa = 3.75MSAATDD209 pKa = 3.29RR210 pKa = 11.84LDD212 pKa = 3.33FQKK215 pKa = 10.84FINEE219 pKa = 3.95YY220 pKa = 9.53FVNISDD226 pKa = 3.74EE227 pKa = 4.17RR228 pKa = 11.84HH229 pKa = 5.83PLLSRR234 pKa = 11.84SIEE237 pKa = 4.13LFKK240 pKa = 10.79DD241 pKa = 3.51TSLSPWSLPGFVADD255 pKa = 4.02MLQMPSNEE263 pKa = 4.03ISWKK267 pKa = 9.82IGQPLGLRR275 pKa = 11.84PSFPLLTIMNCAMASQAVRR294 pKa = 11.84NLDD297 pKa = 3.32GYY299 pKa = 9.15GTYY302 pKa = 8.93NTFACVGDD310 pKa = 4.05DD311 pKa = 4.14LVIQAEE317 pKa = 4.23YY318 pKa = 11.0ADD320 pKa = 4.99AYY322 pKa = 8.82VQVVSEE328 pKa = 4.37FSGLINRR335 pKa = 11.84DD336 pKa = 2.98KK337 pKa = 11.6GMTSSKK343 pKa = 10.32YY344 pKa = 10.79AEE346 pKa = 4.51FCSQLITADD355 pKa = 2.9SSYY358 pKa = 11.38ALKK361 pKa = 10.37PRR363 pKa = 11.84WIDD366 pKa = 3.61SIEE369 pKa = 4.02GSLNNIEE376 pKa = 4.5KK377 pKa = 10.11FQTSGLHH384 pKa = 5.53PKK386 pKa = 9.52VPEE389 pKa = 4.56WIQRR393 pKa = 11.84LYY395 pKa = 10.92NAIAKK400 pKa = 10.04RR401 pKa = 11.84SIKK404 pKa = 10.64GFTKK408 pKa = 10.53YY409 pKa = 10.79GYY411 pKa = 8.5TKK413 pKa = 9.78TDD415 pKa = 3.73CYY417 pKa = 11.48DD418 pKa = 3.42SLKK421 pKa = 11.04DD422 pKa = 3.36RR423 pKa = 11.84VAIHH427 pKa = 7.15DD428 pKa = 4.85FIQSTRR434 pKa = 11.84KK435 pKa = 9.38APRR438 pKa = 11.84DD439 pKa = 3.33IEE441 pKa = 4.79EE442 pKa = 4.01ITLQTLYY449 pKa = 9.94MRR451 pKa = 11.84AVSEE455 pKa = 4.32QDD457 pKa = 3.2EE458 pKa = 4.63LNQKK462 pKa = 8.05VKK464 pKa = 10.84CFDD467 pKa = 4.8LSTSWAMPKK476 pKa = 10.36LGQNKK481 pKa = 8.6QEE483 pKa = 3.54ISTRR487 pKa = 11.84HH488 pKa = 5.62KK489 pKa = 11.03DD490 pKa = 2.94IVQKK494 pKa = 11.06YY495 pKa = 9.57DD496 pKa = 3.16IEE498 pKa = 6.22LGDD501 pKa = 4.35LPSDD505 pKa = 3.52RR506 pKa = 11.84ATSVAVPGIQQWNYY520 pKa = 10.97KK521 pKa = 8.18EE522 pKa = 5.08DD523 pKa = 3.92CYY525 pKa = 11.3SPKK528 pKa = 10.19VSEE531 pKa = 4.71LNTAKK536 pKa = 10.84KK537 pKa = 10.02LLKK540 pKa = 9.46TIEE543 pKa = 4.33SIKK546 pKa = 10.57VEE548 pKa = 4.15SAGTLISASQEE559 pKa = 4.03TKK561 pKa = 9.89HH562 pKa = 6.31CRR564 pKa = 11.84TEE566 pKa = 3.64ILTDD570 pKa = 4.06LSNHH574 pKa = 5.96EE575 pKa = 4.36SVIVHH580 pKa = 5.85TNKK583 pKa = 9.09ATNTTAIQTKK593 pKa = 9.81HH594 pKa = 4.41NQKK597 pKa = 10.34SKK599 pKa = 10.76RR600 pKa = 11.84LEE602 pKa = 4.12DD603 pKa = 3.54LLANLKK609 pKa = 9.93IDD611 pKa = 4.47DD612 pKa = 4.63PTDD615 pKa = 3.35EE616 pKa = 5.11AQTSMEE622 pKa = 3.87TSYY625 pKa = 11.91
Molecular weight: 70.68 kDa
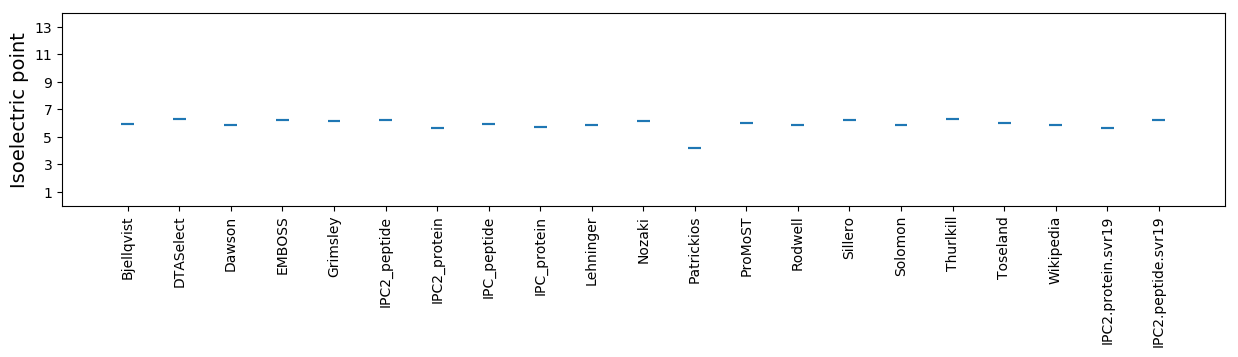
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIM3|A0A1L3KIM3_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 22 OX=1922953 PE=4 SV=1
MM1 pKa = 7.58EE2 pKa = 6.53KK3 pKa = 10.29DD4 pKa = 5.69LKK6 pKa = 10.76MKK8 pKa = 8.65WQQRR12 pKa = 11.84EE13 pKa = 3.99WFNPSHH19 pKa = 6.25LTGSSIPGSGKK30 pKa = 10.49AMSIRR35 pKa = 11.84NRR37 pKa = 11.84NGNISYY43 pKa = 10.95ASIVSAYY50 pKa = 10.42SFSLSTAPLFIWKK63 pKa = 9.46FLDD66 pKa = 4.36DD67 pKa = 3.49IKK69 pKa = 10.87YY70 pKa = 9.71PEE72 pKa = 4.25MVYY75 pKa = 10.18EE76 pKa = 4.07VQGNLEE82 pKa = 3.94QSIKK86 pKa = 10.3LSQIIEE92 pKa = 3.9EE93 pKa = 4.09RR94 pKa = 11.84LQNEE98 pKa = 4.15RR99 pKa = 11.84LLDD102 pKa = 3.84EE103 pKa = 4.98ALSGNNFLDD112 pKa = 4.36DD113 pKa = 4.69DD114 pKa = 4.92SFVDD118 pKa = 3.58YY119 pKa = 11.15QKK121 pKa = 10.93FVTPILGLHH130 pKa = 6.28HH131 pKa = 6.75SVGNIGFLQQGSGKK145 pKa = 9.72LRR147 pKa = 11.84AVANPNRR154 pKa = 11.84FVQYY158 pKa = 10.26CNSPLGEE165 pKa = 4.24VLSSHH170 pKa = 5.58QQGLDD175 pKa = 2.93GCFVYY180 pKa = 10.64NQEE183 pKa = 5.88AGIEE187 pKa = 4.13WVKK190 pKa = 11.01RR191 pKa = 11.84KK192 pKa = 9.74LKK194 pKa = 10.35QGNTLASFDD203 pKa = 3.75MSAATDD209 pKa = 3.29RR210 pKa = 11.84LDD212 pKa = 3.33FQKK215 pKa = 10.84FINEE219 pKa = 3.95YY220 pKa = 9.53FVNISDD226 pKa = 3.74EE227 pKa = 4.17RR228 pKa = 11.84HH229 pKa = 5.83PLLSRR234 pKa = 11.84SIEE237 pKa = 4.13LFKK240 pKa = 10.79DD241 pKa = 3.51TSLSPWSLPGFVADD255 pKa = 4.02MLQMPSNEE263 pKa = 4.03ISWKK267 pKa = 9.82IGQPLGLRR275 pKa = 11.84PSFPLLTIMNCAMASQAVRR294 pKa = 11.84NLDD297 pKa = 3.32GYY299 pKa = 9.15GTYY302 pKa = 8.93NTFACVGDD310 pKa = 4.05DD311 pKa = 4.14LVIQAEE317 pKa = 4.23YY318 pKa = 11.0ADD320 pKa = 4.99AYY322 pKa = 8.82VQVVSEE328 pKa = 4.37FSGLINRR335 pKa = 11.84DD336 pKa = 2.98KK337 pKa = 11.6GMTSSKK343 pKa = 10.32YY344 pKa = 10.79AEE346 pKa = 4.51FCSQLITADD355 pKa = 2.9SSYY358 pKa = 11.38ALKK361 pKa = 10.37PRR363 pKa = 11.84WIDD366 pKa = 3.61SIEE369 pKa = 4.02GSLNNIEE376 pKa = 4.5KK377 pKa = 10.11FQTSGLHH384 pKa = 5.53PKK386 pKa = 9.52VPEE389 pKa = 4.56WIQRR393 pKa = 11.84LYY395 pKa = 10.92NAIAKK400 pKa = 10.04RR401 pKa = 11.84SIKK404 pKa = 10.64GFTKK408 pKa = 10.53YY409 pKa = 10.79GYY411 pKa = 8.5TKK413 pKa = 9.78TDD415 pKa = 3.73CYY417 pKa = 11.48DD418 pKa = 3.42SLKK421 pKa = 11.04DD422 pKa = 3.36RR423 pKa = 11.84VAIHH427 pKa = 7.15DD428 pKa = 4.85FIQSTRR434 pKa = 11.84KK435 pKa = 9.38APRR438 pKa = 11.84DD439 pKa = 3.33IEE441 pKa = 4.79EE442 pKa = 4.01ITLQTLYY449 pKa = 9.94MRR451 pKa = 11.84AVSEE455 pKa = 4.32QDD457 pKa = 3.2EE458 pKa = 4.63LNQKK462 pKa = 8.05VKK464 pKa = 10.84CFDD467 pKa = 4.8LSTSWAMPKK476 pKa = 10.36LGQNKK481 pKa = 8.6QEE483 pKa = 3.54ISTRR487 pKa = 11.84HH488 pKa = 5.62KK489 pKa = 11.03DD490 pKa = 2.94IVQKK494 pKa = 11.06YY495 pKa = 9.57DD496 pKa = 3.16IEE498 pKa = 6.22LGDD501 pKa = 4.35LPSDD505 pKa = 3.52RR506 pKa = 11.84ATSVAVPGIQQWNYY520 pKa = 10.97KK521 pKa = 8.18EE522 pKa = 5.08DD523 pKa = 3.92CYY525 pKa = 11.3SPKK528 pKa = 10.19VSEE531 pKa = 4.71LNTAKK536 pKa = 10.84KK537 pKa = 10.02LLKK540 pKa = 9.46TIEE543 pKa = 4.33SIKK546 pKa = 10.57VEE548 pKa = 4.15SAGTLISASQEE559 pKa = 4.03TKK561 pKa = 9.89HH562 pKa = 6.31CRR564 pKa = 11.84TEE566 pKa = 3.64ILTDD570 pKa = 4.06LSNHH574 pKa = 5.96EE575 pKa = 4.36SVIVHH580 pKa = 5.85TNKK583 pKa = 9.09ATNTTAIQTKK593 pKa = 9.81HH594 pKa = 4.41NQKK597 pKa = 10.34SKK599 pKa = 10.76RR600 pKa = 11.84LEE602 pKa = 4.12DD603 pKa = 3.54LLANLKK609 pKa = 9.93IDD611 pKa = 4.47DD612 pKa = 4.63PTDD615 pKa = 3.35EE616 pKa = 5.11AQTSMEE622 pKa = 3.87TSYY625 pKa = 11.91
MM1 pKa = 7.58EE2 pKa = 6.53KK3 pKa = 10.29DD4 pKa = 5.69LKK6 pKa = 10.76MKK8 pKa = 8.65WQQRR12 pKa = 11.84EE13 pKa = 3.99WFNPSHH19 pKa = 6.25LTGSSIPGSGKK30 pKa = 10.49AMSIRR35 pKa = 11.84NRR37 pKa = 11.84NGNISYY43 pKa = 10.95ASIVSAYY50 pKa = 10.42SFSLSTAPLFIWKK63 pKa = 9.46FLDD66 pKa = 4.36DD67 pKa = 3.49IKK69 pKa = 10.87YY70 pKa = 9.71PEE72 pKa = 4.25MVYY75 pKa = 10.18EE76 pKa = 4.07VQGNLEE82 pKa = 3.94QSIKK86 pKa = 10.3LSQIIEE92 pKa = 3.9EE93 pKa = 4.09RR94 pKa = 11.84LQNEE98 pKa = 4.15RR99 pKa = 11.84LLDD102 pKa = 3.84EE103 pKa = 4.98ALSGNNFLDD112 pKa = 4.36DD113 pKa = 4.69DD114 pKa = 4.92SFVDD118 pKa = 3.58YY119 pKa = 11.15QKK121 pKa = 10.93FVTPILGLHH130 pKa = 6.28HH131 pKa = 6.75SVGNIGFLQQGSGKK145 pKa = 9.72LRR147 pKa = 11.84AVANPNRR154 pKa = 11.84FVQYY158 pKa = 10.26CNSPLGEE165 pKa = 4.24VLSSHH170 pKa = 5.58QQGLDD175 pKa = 2.93GCFVYY180 pKa = 10.64NQEE183 pKa = 5.88AGIEE187 pKa = 4.13WVKK190 pKa = 11.01RR191 pKa = 11.84KK192 pKa = 9.74LKK194 pKa = 10.35QGNTLASFDD203 pKa = 3.75MSAATDD209 pKa = 3.29RR210 pKa = 11.84LDD212 pKa = 3.33FQKK215 pKa = 10.84FINEE219 pKa = 3.95YY220 pKa = 9.53FVNISDD226 pKa = 3.74EE227 pKa = 4.17RR228 pKa = 11.84HH229 pKa = 5.83PLLSRR234 pKa = 11.84SIEE237 pKa = 4.13LFKK240 pKa = 10.79DD241 pKa = 3.51TSLSPWSLPGFVADD255 pKa = 4.02MLQMPSNEE263 pKa = 4.03ISWKK267 pKa = 9.82IGQPLGLRR275 pKa = 11.84PSFPLLTIMNCAMASQAVRR294 pKa = 11.84NLDD297 pKa = 3.32GYY299 pKa = 9.15GTYY302 pKa = 8.93NTFACVGDD310 pKa = 4.05DD311 pKa = 4.14LVIQAEE317 pKa = 4.23YY318 pKa = 11.0ADD320 pKa = 4.99AYY322 pKa = 8.82VQVVSEE328 pKa = 4.37FSGLINRR335 pKa = 11.84DD336 pKa = 2.98KK337 pKa = 11.6GMTSSKK343 pKa = 10.32YY344 pKa = 10.79AEE346 pKa = 4.51FCSQLITADD355 pKa = 2.9SSYY358 pKa = 11.38ALKK361 pKa = 10.37PRR363 pKa = 11.84WIDD366 pKa = 3.61SIEE369 pKa = 4.02GSLNNIEE376 pKa = 4.5KK377 pKa = 10.11FQTSGLHH384 pKa = 5.53PKK386 pKa = 9.52VPEE389 pKa = 4.56WIQRR393 pKa = 11.84LYY395 pKa = 10.92NAIAKK400 pKa = 10.04RR401 pKa = 11.84SIKK404 pKa = 10.64GFTKK408 pKa = 10.53YY409 pKa = 10.79GYY411 pKa = 8.5TKK413 pKa = 9.78TDD415 pKa = 3.73CYY417 pKa = 11.48DD418 pKa = 3.42SLKK421 pKa = 11.04DD422 pKa = 3.36RR423 pKa = 11.84VAIHH427 pKa = 7.15DD428 pKa = 4.85FIQSTRR434 pKa = 11.84KK435 pKa = 9.38APRR438 pKa = 11.84DD439 pKa = 3.33IEE441 pKa = 4.79EE442 pKa = 4.01ITLQTLYY449 pKa = 9.94MRR451 pKa = 11.84AVSEE455 pKa = 4.32QDD457 pKa = 3.2EE458 pKa = 4.63LNQKK462 pKa = 8.05VKK464 pKa = 10.84CFDD467 pKa = 4.8LSTSWAMPKK476 pKa = 10.36LGQNKK481 pKa = 8.6QEE483 pKa = 3.54ISTRR487 pKa = 11.84HH488 pKa = 5.62KK489 pKa = 11.03DD490 pKa = 2.94IVQKK494 pKa = 11.06YY495 pKa = 9.57DD496 pKa = 3.16IEE498 pKa = 6.22LGDD501 pKa = 4.35LPSDD505 pKa = 3.52RR506 pKa = 11.84ATSVAVPGIQQWNYY520 pKa = 10.97KK521 pKa = 8.18EE522 pKa = 5.08DD523 pKa = 3.92CYY525 pKa = 11.3SPKK528 pKa = 10.19VSEE531 pKa = 4.71LNTAKK536 pKa = 10.84KK537 pKa = 10.02LLKK540 pKa = 9.46TIEE543 pKa = 4.33SIKK546 pKa = 10.57VEE548 pKa = 4.15SAGTLISASQEE559 pKa = 4.03TKK561 pKa = 9.89HH562 pKa = 6.31CRR564 pKa = 11.84TEE566 pKa = 3.64ILTDD570 pKa = 4.06LSNHH574 pKa = 5.96EE575 pKa = 4.36SVIVHH580 pKa = 5.85TNKK583 pKa = 9.09ATNTTAIQTKK593 pKa = 9.81HH594 pKa = 4.41NQKK597 pKa = 10.34SKK599 pKa = 10.76RR600 pKa = 11.84LEE602 pKa = 4.12DD603 pKa = 3.54LLANLKK609 pKa = 9.93IDD611 pKa = 4.47DD612 pKa = 4.63PTDD615 pKa = 3.35EE616 pKa = 5.11AQTSMEE622 pKa = 3.87TSYY625 pKa = 11.91
Molecular weight: 70.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
625 |
625 |
625 |
625.0 |
70.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.92 ± 0.0 | 1.44 ± 0.0 |
6.24 ± 0.0 | 6.08 ± 0.0 |
3.84 ± 0.0 | 5.12 ± 0.0 |
1.92 ± 0.0 | 6.72 ± 0.0 |
7.04 ± 0.0 | 9.28 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.08 ± 0.0 | 5.28 ± 0.0 |
3.68 ± 0.0 | 5.76 ± 0.0 |
4.0 ± 0.0 | 9.92 ± 0.0 |
5.6 ± 0.0 | 4.8 ± 0.0 |
1.6 ± 0.0 | 3.68 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
