
Cucurbit aphid-borne yellows virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus
Average proteome isoelectric point is 7.69
Get precalculated fractions of proteins
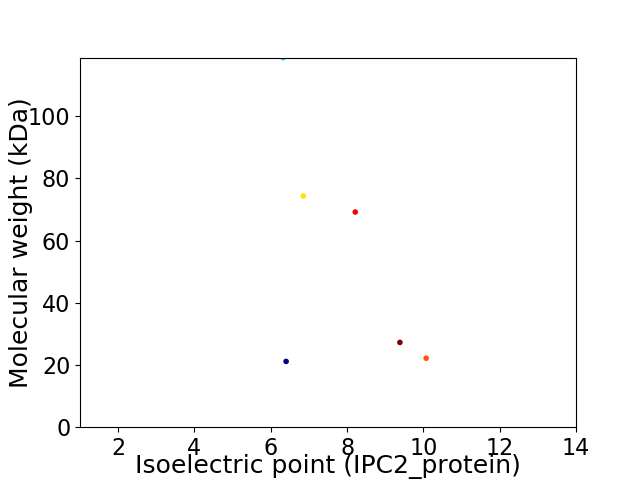
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
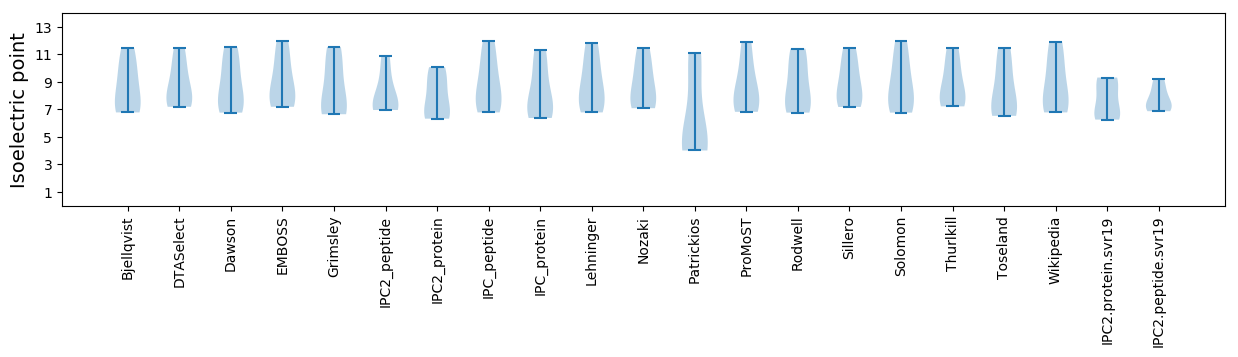
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q65970|Q65970_9LUTE Coat protein OS=Cucurbit aphid-borne yellows virus OX=91753 PE=4 SV=1
MM1 pKa = 7.65APTYY5 pKa = 10.11FAFFLLVCLCSSVSSYY21 pKa = 10.8QGTMFIPLEE30 pKa = 4.16PANASYY36 pKa = 10.32WLDD39 pKa = 3.5STDD42 pKa = 3.09IAVPPSHH49 pKa = 6.92PQVQLIYY56 pKa = 10.38DD57 pKa = 4.44CPPQKK62 pKa = 9.89MLRR65 pKa = 11.84DD66 pKa = 4.32FSSRR70 pKa = 11.84DD71 pKa = 2.89ITLEE75 pKa = 3.56LWEE78 pKa = 4.46RR79 pKa = 11.84GYY81 pKa = 11.46NEE83 pKa = 3.54TRR85 pKa = 11.84QAFSEE90 pKa = 4.3AMQNLQNLLMSGVRR104 pKa = 11.84QFHH107 pKa = 6.35TGLEE111 pKa = 4.29SLLHH115 pKa = 6.44VILQTAAYY123 pKa = 9.07LWTSLIWATACATWYY138 pKa = 10.6LLSKK142 pKa = 9.09YY143 pKa = 8.12TIEE146 pKa = 4.12MLTLASLYY154 pKa = 10.09ISTVYY159 pKa = 8.23MVKK162 pKa = 9.17MAAWIFGDD170 pKa = 4.92LPISLLKK177 pKa = 10.8AGLSMVRR184 pKa = 11.84GVSRR188 pKa = 11.84ALWYY192 pKa = 10.11KK193 pKa = 10.38RR194 pKa = 11.84SYY196 pKa = 10.09NAEE199 pKa = 4.01KK200 pKa = 10.58SVEE203 pKa = 4.2GYY205 pKa = 11.01LSFKK209 pKa = 10.49IPQNPPKK216 pKa = 10.74NSVLQVQYY224 pKa = 11.22KK225 pKa = 9.94DD226 pKa = 3.8GSHH229 pKa = 6.69AGYY232 pKa = 8.07ATCVTLYY239 pKa = 10.85NGTNGLLTAYY249 pKa = 9.58HH250 pKa = 5.73VAVPGSKK257 pKa = 10.08VVSTRR262 pKa = 11.84NGNKK266 pKa = 9.2VPLSEE271 pKa = 4.51FRR273 pKa = 11.84SIMEE277 pKa = 3.91SEE279 pKa = 4.06KK280 pKa = 10.67RR281 pKa = 11.84DD282 pKa = 3.77LVLLAGPPNWEE293 pKa = 3.62GTLACKK299 pKa = 10.06AVQFQSAQNLCKK311 pKa = 10.58SKK313 pKa = 11.09ASFYY317 pKa = 10.84AYY319 pKa = 10.15DD320 pKa = 3.74GEE322 pKa = 4.67GWISSNAEE330 pKa = 3.36IVGIAEE336 pKa = 4.61GKK338 pKa = 7.08THH340 pKa = 7.5ASVLSNTDD348 pKa = 3.14AGHH351 pKa = 6.74SGTPYY356 pKa = 9.83FNGRR360 pKa = 11.84TVLGVHH366 pKa = 6.02VGGAKK371 pKa = 10.31DD372 pKa = 3.51EE373 pKa = 4.43NFNYY377 pKa = 7.68MAPIPPVYY385 pKa = 10.58GLTSPSYY392 pKa = 10.18EE393 pKa = 4.28FEE395 pKa = 3.95TTAPQGRR402 pKa = 11.84LFTQEE407 pKa = 4.23EE408 pKa = 4.28IEE410 pKa = 4.08EE411 pKa = 4.74LIEE414 pKa = 4.0EE415 pKa = 4.57FSFSEE420 pKa = 3.77ITSIMGHH427 pKa = 5.39RR428 pKa = 11.84RR429 pKa = 11.84FHH431 pKa = 6.85QMHH434 pKa = 7.39DD435 pKa = 4.0SQRR438 pKa = 11.84HH439 pKa = 3.72QADD442 pKa = 3.47YY443 pKa = 10.41EE444 pKa = 4.34YY445 pKa = 10.89EE446 pKa = 4.05SGKK449 pKa = 10.33RR450 pKa = 11.84AGGGDD455 pKa = 2.9RR456 pKa = 11.84RR457 pKa = 11.84NNRR460 pKa = 11.84TRR462 pKa = 11.84CHH464 pKa = 5.61PRR466 pKa = 11.84GWAHH470 pKa = 6.14KK471 pKa = 7.71WRR473 pKa = 11.84RR474 pKa = 11.84TVSCCSLFPEE484 pKa = 5.11GQHH487 pKa = 5.2VHH489 pKa = 5.55TSEE492 pKa = 4.13QTGKK496 pKa = 8.44PHH498 pKa = 6.84CFCSIPHH505 pKa = 5.68HH506 pKa = 5.36QCFYY510 pKa = 10.56RR511 pKa = 11.84HH512 pKa = 5.61FVGNQTGNNGQNRR525 pKa = 11.84CPFNRR530 pKa = 11.84EE531 pKa = 3.25ASGPSLGGQSHH542 pKa = 6.87EE543 pKa = 4.34EE544 pKa = 4.12TEE546 pKa = 4.6VAQTRR551 pKa = 11.84QEE553 pKa = 4.11KK554 pKa = 10.41FEE556 pKa = 4.07EE557 pKa = 4.35LATDD561 pKa = 4.34FSSFFDD567 pKa = 3.83AQYY570 pKa = 9.27TWEE573 pKa = 4.22QGGEE577 pKa = 3.98EE578 pKa = 4.35APGFDD583 pKa = 4.49KK584 pKa = 11.31VGSLPQFYY592 pKa = 9.77HH593 pKa = 7.1AKK595 pKa = 9.43QKK597 pKa = 10.8KK598 pKa = 8.26SSNWGDD604 pKa = 4.07KK605 pKa = 9.79ICKK608 pKa = 7.79QHH610 pKa = 7.24PEE612 pKa = 4.09MGDD615 pKa = 3.31LTKK618 pKa = 11.17GFGWPQFGAKK628 pKa = 10.1AEE630 pKa = 4.19LKK632 pKa = 10.47SLRR635 pKa = 11.84LQAARR640 pKa = 11.84WLEE643 pKa = 3.8RR644 pKa = 11.84AQSVKK649 pKa = 10.18IPSSEE654 pKa = 3.78EE655 pKa = 3.63RR656 pKa = 11.84EE657 pKa = 4.21HH658 pKa = 8.71VIEE661 pKa = 4.38RR662 pKa = 11.84CCRR665 pKa = 11.84AYY667 pKa = 10.26QAAKK671 pKa = 9.66TNGPMATRR679 pKa = 11.84GDD681 pKa = 3.71RR682 pKa = 11.84LSWDD686 pKa = 3.35NFLQDD691 pKa = 3.64FKK693 pKa = 11.4QAVLSLEE700 pKa = 3.82MDD702 pKa = 3.6AGIGVPYY709 pKa = 9.92IAYY712 pKa = 8.45GKK714 pKa = 6.46PTHH717 pKa = 6.59RR718 pKa = 11.84GWVEE722 pKa = 3.91DD723 pKa = 3.97KK724 pKa = 11.04KK725 pKa = 11.11LLPILARR732 pKa = 11.84LTFNRR737 pKa = 11.84LQKK740 pKa = 9.25MLEE743 pKa = 3.77VRR745 pKa = 11.84YY746 pKa = 10.3VDD748 pKa = 4.19LSPAEE753 pKa = 3.89LVRR756 pKa = 11.84RR757 pKa = 11.84GLCDD761 pKa = 4.7PIRR764 pKa = 11.84VFVKK768 pKa = 10.82GEE770 pKa = 3.63PHH772 pKa = 6.0KK773 pKa = 10.35QAKK776 pKa = 9.74LDD778 pKa = 3.49EE779 pKa = 4.35GRR781 pKa = 11.84YY782 pKa = 8.74RR783 pKa = 11.84LIMSVSLVDD792 pKa = 3.45QLVARR797 pKa = 11.84VLFQNQNKK805 pKa = 9.66RR806 pKa = 11.84EE807 pKa = 3.81IALWRR812 pKa = 11.84VVPSKK817 pKa = 10.72PGFGLSTDD825 pKa = 3.58EE826 pKa = 4.04QVAEE830 pKa = 4.3FMQILSAQVGLTPSDD845 pKa = 5.62LITEE849 pKa = 4.69WRR851 pKa = 11.84AHH853 pKa = 6.44MIATDD858 pKa = 3.65CSGFDD863 pKa = 3.32WSVSDD868 pKa = 4.58WLLEE872 pKa = 4.07DD873 pKa = 4.28DD874 pKa = 3.82MEE876 pKa = 4.4VRR878 pKa = 11.84NRR880 pKa = 11.84LTLDD884 pKa = 3.27LNEE887 pKa = 3.95TTRR890 pKa = 11.84RR891 pKa = 11.84LRR893 pKa = 11.84SWLYY897 pKa = 10.5CISNSVLCLSDD908 pKa = 3.31GTLLAQRR915 pKa = 11.84VPGVQKK921 pKa = 10.45SGSYY925 pKa = 8.25NTSSSNSRR933 pKa = 11.84IRR935 pKa = 11.84VMAAYY940 pKa = 9.93HH941 pKa = 6.65CGAEE945 pKa = 3.89WAMAMGDD952 pKa = 3.92DD953 pKa = 4.41ALEE956 pKa = 4.35SACSNLEE963 pKa = 3.95RR964 pKa = 11.84YY965 pKa = 9.28KK966 pKa = 10.97SLGFKK971 pKa = 10.48VEE973 pKa = 4.14EE974 pKa = 4.08SSKK977 pKa = 11.47LEE979 pKa = 3.98FCSHH983 pKa = 6.1IFEE986 pKa = 5.2KK987 pKa = 10.64EE988 pKa = 3.74DD989 pKa = 3.82LAIPVNKK996 pKa = 10.52AKK998 pKa = 9.87MLYY1001 pKa = 10.13KK1002 pKa = 10.31LIHH1005 pKa = 6.42GYY1007 pKa = 9.43EE1008 pKa = 4.18PEE1010 pKa = 4.59CGNVEE1015 pKa = 4.12VLINYY1020 pKa = 7.74LAACFSILNEE1030 pKa = 3.97LRR1032 pKa = 11.84SDD1034 pKa = 3.67PSLVEE1039 pKa = 4.43TLHH1042 pKa = 5.54QWLVAPVQPQKK1053 pKa = 10.84II1054 pKa = 3.87
MM1 pKa = 7.65APTYY5 pKa = 10.11FAFFLLVCLCSSVSSYY21 pKa = 10.8QGTMFIPLEE30 pKa = 4.16PANASYY36 pKa = 10.32WLDD39 pKa = 3.5STDD42 pKa = 3.09IAVPPSHH49 pKa = 6.92PQVQLIYY56 pKa = 10.38DD57 pKa = 4.44CPPQKK62 pKa = 9.89MLRR65 pKa = 11.84DD66 pKa = 4.32FSSRR70 pKa = 11.84DD71 pKa = 2.89ITLEE75 pKa = 3.56LWEE78 pKa = 4.46RR79 pKa = 11.84GYY81 pKa = 11.46NEE83 pKa = 3.54TRR85 pKa = 11.84QAFSEE90 pKa = 4.3AMQNLQNLLMSGVRR104 pKa = 11.84QFHH107 pKa = 6.35TGLEE111 pKa = 4.29SLLHH115 pKa = 6.44VILQTAAYY123 pKa = 9.07LWTSLIWATACATWYY138 pKa = 10.6LLSKK142 pKa = 9.09YY143 pKa = 8.12TIEE146 pKa = 4.12MLTLASLYY154 pKa = 10.09ISTVYY159 pKa = 8.23MVKK162 pKa = 9.17MAAWIFGDD170 pKa = 4.92LPISLLKK177 pKa = 10.8AGLSMVRR184 pKa = 11.84GVSRR188 pKa = 11.84ALWYY192 pKa = 10.11KK193 pKa = 10.38RR194 pKa = 11.84SYY196 pKa = 10.09NAEE199 pKa = 4.01KK200 pKa = 10.58SVEE203 pKa = 4.2GYY205 pKa = 11.01LSFKK209 pKa = 10.49IPQNPPKK216 pKa = 10.74NSVLQVQYY224 pKa = 11.22KK225 pKa = 9.94DD226 pKa = 3.8GSHH229 pKa = 6.69AGYY232 pKa = 8.07ATCVTLYY239 pKa = 10.85NGTNGLLTAYY249 pKa = 9.58HH250 pKa = 5.73VAVPGSKK257 pKa = 10.08VVSTRR262 pKa = 11.84NGNKK266 pKa = 9.2VPLSEE271 pKa = 4.51FRR273 pKa = 11.84SIMEE277 pKa = 3.91SEE279 pKa = 4.06KK280 pKa = 10.67RR281 pKa = 11.84DD282 pKa = 3.77LVLLAGPPNWEE293 pKa = 3.62GTLACKK299 pKa = 10.06AVQFQSAQNLCKK311 pKa = 10.58SKK313 pKa = 11.09ASFYY317 pKa = 10.84AYY319 pKa = 10.15DD320 pKa = 3.74GEE322 pKa = 4.67GWISSNAEE330 pKa = 3.36IVGIAEE336 pKa = 4.61GKK338 pKa = 7.08THH340 pKa = 7.5ASVLSNTDD348 pKa = 3.14AGHH351 pKa = 6.74SGTPYY356 pKa = 9.83FNGRR360 pKa = 11.84TVLGVHH366 pKa = 6.02VGGAKK371 pKa = 10.31DD372 pKa = 3.51EE373 pKa = 4.43NFNYY377 pKa = 7.68MAPIPPVYY385 pKa = 10.58GLTSPSYY392 pKa = 10.18EE393 pKa = 4.28FEE395 pKa = 3.95TTAPQGRR402 pKa = 11.84LFTQEE407 pKa = 4.23EE408 pKa = 4.28IEE410 pKa = 4.08EE411 pKa = 4.74LIEE414 pKa = 4.0EE415 pKa = 4.57FSFSEE420 pKa = 3.77ITSIMGHH427 pKa = 5.39RR428 pKa = 11.84RR429 pKa = 11.84FHH431 pKa = 6.85QMHH434 pKa = 7.39DD435 pKa = 4.0SQRR438 pKa = 11.84HH439 pKa = 3.72QADD442 pKa = 3.47YY443 pKa = 10.41EE444 pKa = 4.34YY445 pKa = 10.89EE446 pKa = 4.05SGKK449 pKa = 10.33RR450 pKa = 11.84AGGGDD455 pKa = 2.9RR456 pKa = 11.84RR457 pKa = 11.84NNRR460 pKa = 11.84TRR462 pKa = 11.84CHH464 pKa = 5.61PRR466 pKa = 11.84GWAHH470 pKa = 6.14KK471 pKa = 7.71WRR473 pKa = 11.84RR474 pKa = 11.84TVSCCSLFPEE484 pKa = 5.11GQHH487 pKa = 5.2VHH489 pKa = 5.55TSEE492 pKa = 4.13QTGKK496 pKa = 8.44PHH498 pKa = 6.84CFCSIPHH505 pKa = 5.68HH506 pKa = 5.36QCFYY510 pKa = 10.56RR511 pKa = 11.84HH512 pKa = 5.61FVGNQTGNNGQNRR525 pKa = 11.84CPFNRR530 pKa = 11.84EE531 pKa = 3.25ASGPSLGGQSHH542 pKa = 6.87EE543 pKa = 4.34EE544 pKa = 4.12TEE546 pKa = 4.6VAQTRR551 pKa = 11.84QEE553 pKa = 4.11KK554 pKa = 10.41FEE556 pKa = 4.07EE557 pKa = 4.35LATDD561 pKa = 4.34FSSFFDD567 pKa = 3.83AQYY570 pKa = 9.27TWEE573 pKa = 4.22QGGEE577 pKa = 3.98EE578 pKa = 4.35APGFDD583 pKa = 4.49KK584 pKa = 11.31VGSLPQFYY592 pKa = 9.77HH593 pKa = 7.1AKK595 pKa = 9.43QKK597 pKa = 10.8KK598 pKa = 8.26SSNWGDD604 pKa = 4.07KK605 pKa = 9.79ICKK608 pKa = 7.79QHH610 pKa = 7.24PEE612 pKa = 4.09MGDD615 pKa = 3.31LTKK618 pKa = 11.17GFGWPQFGAKK628 pKa = 10.1AEE630 pKa = 4.19LKK632 pKa = 10.47SLRR635 pKa = 11.84LQAARR640 pKa = 11.84WLEE643 pKa = 3.8RR644 pKa = 11.84AQSVKK649 pKa = 10.18IPSSEE654 pKa = 3.78EE655 pKa = 3.63RR656 pKa = 11.84EE657 pKa = 4.21HH658 pKa = 8.71VIEE661 pKa = 4.38RR662 pKa = 11.84CCRR665 pKa = 11.84AYY667 pKa = 10.26QAAKK671 pKa = 9.66TNGPMATRR679 pKa = 11.84GDD681 pKa = 3.71RR682 pKa = 11.84LSWDD686 pKa = 3.35NFLQDD691 pKa = 3.64FKK693 pKa = 11.4QAVLSLEE700 pKa = 3.82MDD702 pKa = 3.6AGIGVPYY709 pKa = 9.92IAYY712 pKa = 8.45GKK714 pKa = 6.46PTHH717 pKa = 6.59RR718 pKa = 11.84GWVEE722 pKa = 3.91DD723 pKa = 3.97KK724 pKa = 11.04KK725 pKa = 11.11LLPILARR732 pKa = 11.84LTFNRR737 pKa = 11.84LQKK740 pKa = 9.25MLEE743 pKa = 3.77VRR745 pKa = 11.84YY746 pKa = 10.3VDD748 pKa = 4.19LSPAEE753 pKa = 3.89LVRR756 pKa = 11.84RR757 pKa = 11.84GLCDD761 pKa = 4.7PIRR764 pKa = 11.84VFVKK768 pKa = 10.82GEE770 pKa = 3.63PHH772 pKa = 6.0KK773 pKa = 10.35QAKK776 pKa = 9.74LDD778 pKa = 3.49EE779 pKa = 4.35GRR781 pKa = 11.84YY782 pKa = 8.74RR783 pKa = 11.84LIMSVSLVDD792 pKa = 3.45QLVARR797 pKa = 11.84VLFQNQNKK805 pKa = 9.66RR806 pKa = 11.84EE807 pKa = 3.81IALWRR812 pKa = 11.84VVPSKK817 pKa = 10.72PGFGLSTDD825 pKa = 3.58EE826 pKa = 4.04QVAEE830 pKa = 4.3FMQILSAQVGLTPSDD845 pKa = 5.62LITEE849 pKa = 4.69WRR851 pKa = 11.84AHH853 pKa = 6.44MIATDD858 pKa = 3.65CSGFDD863 pKa = 3.32WSVSDD868 pKa = 4.58WLLEE872 pKa = 4.07DD873 pKa = 4.28DD874 pKa = 3.82MEE876 pKa = 4.4VRR878 pKa = 11.84NRR880 pKa = 11.84LTLDD884 pKa = 3.27LNEE887 pKa = 3.95TTRR890 pKa = 11.84RR891 pKa = 11.84LRR893 pKa = 11.84SWLYY897 pKa = 10.5CISNSVLCLSDD908 pKa = 3.31GTLLAQRR915 pKa = 11.84VPGVQKK921 pKa = 10.45SGSYY925 pKa = 8.25NTSSSNSRR933 pKa = 11.84IRR935 pKa = 11.84VMAAYY940 pKa = 9.93HH941 pKa = 6.65CGAEE945 pKa = 3.89WAMAMGDD952 pKa = 3.92DD953 pKa = 4.41ALEE956 pKa = 4.35SACSNLEE963 pKa = 3.95RR964 pKa = 11.84YY965 pKa = 9.28KK966 pKa = 10.97SLGFKK971 pKa = 10.48VEE973 pKa = 4.14EE974 pKa = 4.08SSKK977 pKa = 11.47LEE979 pKa = 3.98FCSHH983 pKa = 6.1IFEE986 pKa = 5.2KK987 pKa = 10.64EE988 pKa = 3.74DD989 pKa = 3.82LAIPVNKK996 pKa = 10.52AKK998 pKa = 9.87MLYY1001 pKa = 10.13KK1002 pKa = 10.31LIHH1005 pKa = 6.42GYY1007 pKa = 9.43EE1008 pKa = 4.18PEE1010 pKa = 4.59CGNVEE1015 pKa = 4.12VLINYY1020 pKa = 7.74LAACFSILNEE1030 pKa = 3.97LRR1032 pKa = 11.84SDD1034 pKa = 3.67PSLVEE1039 pKa = 4.43TLHH1042 pKa = 5.54QWLVAPVQPQKK1053 pKa = 10.84II1054 pKa = 3.87
Molecular weight: 118.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q65968|Q65968_9LUTE Serine protease OS=Cucurbit aphid-borne yellows virus OX=91753 PE=4 SV=1
MM1 pKa = 7.29QIEE4 pKa = 4.56SVKK7 pKa = 10.07QQLIFRR13 pKa = 11.84PTRR16 pKa = 11.84RR17 pKa = 11.84TSINDD22 pKa = 3.15RR23 pKa = 11.84KK24 pKa = 10.88LNVANFLINHH34 pKa = 5.98SFFLAINGTNLLRR47 pKa = 11.84LFLARR52 pKa = 11.84LPLLISEE59 pKa = 4.56QLSGDD64 pKa = 3.65YY65 pKa = 10.69VYY67 pKa = 10.95TPGASKK73 pKa = 10.7RR74 pKa = 11.84IILARR79 pKa = 11.84FHH81 pKa = 6.21RR82 pKa = 11.84HH83 pKa = 5.68CGAPLPSSSAVDD95 pKa = 3.7LRR97 pKa = 11.84LPASKK102 pKa = 10.55DD103 pKa = 3.19VARR106 pKa = 11.84FFLARR111 pKa = 11.84HH112 pKa = 5.27YY113 pKa = 11.19SRR115 pKa = 11.84ALGEE119 pKa = 4.17RR120 pKa = 11.84IQRR123 pKa = 11.84NQTSLFRR130 pKa = 11.84GYY132 pKa = 11.46AEE134 pKa = 3.67FAKK137 pKa = 10.27FINVWCSSISHH148 pKa = 6.93RR149 pKa = 11.84LGEE152 pKa = 3.99FTPRR156 pKa = 11.84NFTNGSIFVDD166 pKa = 4.63LSNLGHH172 pKa = 5.94SLCDD176 pKa = 3.28LVLAQQVHH184 pKa = 6.8DD185 pKa = 3.86RR186 pKa = 11.84DD187 pKa = 3.81AYY189 pKa = 9.56TRR191 pKa = 11.84LALHH195 pKa = 6.09IHH197 pKa = 7.1RR198 pKa = 11.84IYY200 pKa = 11.26GEE202 pKa = 4.34DD203 pKa = 3.29GGLDD207 pKa = 3.71FWRR210 pKa = 11.84LANFPSKK217 pKa = 10.14SWPFNGEE224 pKa = 3.59RR225 pKa = 11.84CLEE228 pKa = 4.07GSVVQKK234 pKa = 10.44EE235 pKa = 4.25LQRR238 pKa = 4.96
MM1 pKa = 7.29QIEE4 pKa = 4.56SVKK7 pKa = 10.07QQLIFRR13 pKa = 11.84PTRR16 pKa = 11.84RR17 pKa = 11.84TSINDD22 pKa = 3.15RR23 pKa = 11.84KK24 pKa = 10.88LNVANFLINHH34 pKa = 5.98SFFLAINGTNLLRR47 pKa = 11.84LFLARR52 pKa = 11.84LPLLISEE59 pKa = 4.56QLSGDD64 pKa = 3.65YY65 pKa = 10.69VYY67 pKa = 10.95TPGASKK73 pKa = 10.7RR74 pKa = 11.84IILARR79 pKa = 11.84FHH81 pKa = 6.21RR82 pKa = 11.84HH83 pKa = 5.68CGAPLPSSSAVDD95 pKa = 3.7LRR97 pKa = 11.84LPASKK102 pKa = 10.55DD103 pKa = 3.19VARR106 pKa = 11.84FFLARR111 pKa = 11.84HH112 pKa = 5.27YY113 pKa = 11.19SRR115 pKa = 11.84ALGEE119 pKa = 4.17RR120 pKa = 11.84IQRR123 pKa = 11.84NQTSLFRR130 pKa = 11.84GYY132 pKa = 11.46AEE134 pKa = 3.67FAKK137 pKa = 10.27FINVWCSSISHH148 pKa = 6.93RR149 pKa = 11.84LGEE152 pKa = 3.99FTPRR156 pKa = 11.84NFTNGSIFVDD166 pKa = 4.63LSNLGHH172 pKa = 5.94SLCDD176 pKa = 3.28LVLAQQVHH184 pKa = 6.8DD185 pKa = 3.86RR186 pKa = 11.84DD187 pKa = 3.81AYY189 pKa = 9.56TRR191 pKa = 11.84LALHH195 pKa = 6.09IHH197 pKa = 7.1RR198 pKa = 11.84IYY200 pKa = 11.26GEE202 pKa = 4.34DD203 pKa = 3.29GGLDD207 pKa = 3.71FWRR210 pKa = 11.84LANFPSKK217 pKa = 10.14SWPFNGEE224 pKa = 3.59RR225 pKa = 11.84CLEE228 pKa = 4.07GSVVQKK234 pKa = 10.44EE235 pKa = 4.25LQRR238 pKa = 4.96
Molecular weight: 27.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2979 |
191 |
1054 |
496.5 |
55.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.486 ± 0.473 | 1.578 ± 0.367 |
4.23 ± 0.571 | 6.244 ± 0.657 |
3.693 ± 0.484 | 6.949 ± 0.418 |
2.216 ± 0.387 | 4.33 ± 0.37 |
4.901 ± 0.425 | 8.191 ± 0.902 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.048 ± 0.286 | 4.532 ± 0.373 |
5.74 ± 0.566 | 4.498 ± 0.253 |
6.949 ± 1.058 | 10.305 ± 0.799 |
5.505 ± 0.507 | 5.27 ± 0.367 |
1.981 ± 0.244 | 3.357 ± 0.342 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
