
Lake Sarah-associated circular virus-41
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.83
Get precalculated fractions of proteins
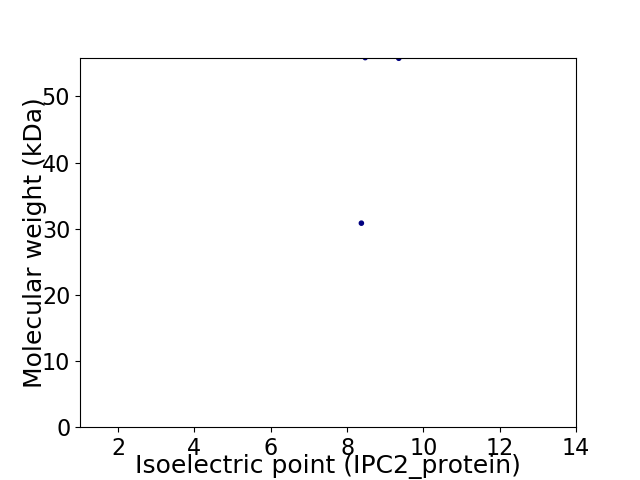
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GA81|A0A126GA81_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-41 OX=1685770 PE=4 SV=1
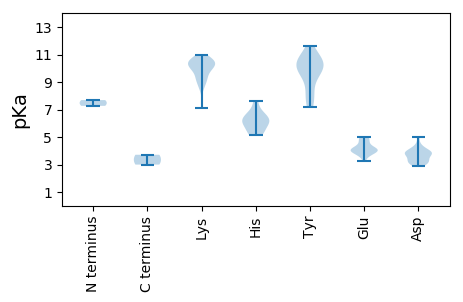
MM1 pKa = 7.69ANGKK5 pKa = 7.14STRR8 pKa = 11.84RR9 pKa = 11.84QGIFWLCTIPSPNEE23 pKa = 3.27VCAQLEE29 pKa = 4.56LGTLPVGLVWCKK41 pKa = 9.04GQKK44 pKa = 9.33EE45 pKa = 4.23RR46 pKa = 11.84GSGTGYY52 pKa = 10.07EE53 pKa = 4.47HH54 pKa = 6.47YY55 pKa = 10.8QLVAAFAKK63 pKa = 10.08KK64 pKa = 10.04VSLPGVVGMFGRR76 pKa = 11.84GTHH79 pKa = 6.3GEE81 pKa = 4.04LSRR84 pKa = 11.84SEE86 pKa = 4.07AANEE90 pKa = 3.88YY91 pKa = 9.79VGKK94 pKa = 10.34EE95 pKa = 3.75EE96 pKa = 4.11TRR98 pKa = 11.84IGTTFEE104 pKa = 4.97LGAKK108 pKa = 9.57PIRR111 pKa = 11.84RR112 pKa = 11.84NSKK115 pKa = 8.97TDD117 pKa = 3.12WEE119 pKa = 4.97SVWTAAKK126 pKa = 10.54SGDD129 pKa = 3.9LSAVPANVRR138 pKa = 11.84VVNYY142 pKa = 9.29HH143 pKa = 6.17ALRR146 pKa = 11.84AIRR149 pKa = 11.84SDD151 pKa = 3.1HH152 pKa = 5.96AVPVGVEE159 pKa = 3.97RR160 pKa = 11.84TVHH163 pKa = 5.15VFWGSTGTGKK173 pKa = 10.02SRR175 pKa = 11.84RR176 pKa = 11.84AWEE179 pKa = 3.79EE180 pKa = 3.55AGIDD184 pKa = 4.16AYY186 pKa = 8.93TKK188 pKa = 10.48CPRR191 pKa = 11.84SKK193 pKa = 10.48FWDD196 pKa = 4.98GYY198 pKa = 10.19QDD200 pKa = 3.52QQHH203 pKa = 5.16VVVDD207 pKa = 4.2EE208 pKa = 4.15FRR210 pKa = 11.84GGIDD214 pKa = 3.25VAHH217 pKa = 7.59LLRR220 pKa = 11.84WFDD223 pKa = 3.83RR224 pKa = 11.84YY225 pKa = 9.38PVRR228 pKa = 11.84VEE230 pKa = 3.72IKK232 pKa = 9.65GSSRR236 pKa = 11.84PLVANTIWITSNKK249 pKa = 10.05DD250 pKa = 3.12PVEE253 pKa = 4.22WYY255 pKa = 9.79PEE257 pKa = 3.88LDD259 pKa = 3.67LATLEE264 pKa = 4.13ALLRR268 pKa = 11.84RR269 pKa = 11.84LTIVHH274 pKa = 6.7FPP276 pKa = 3.71
MM1 pKa = 7.69ANGKK5 pKa = 7.14STRR8 pKa = 11.84RR9 pKa = 11.84QGIFWLCTIPSPNEE23 pKa = 3.27VCAQLEE29 pKa = 4.56LGTLPVGLVWCKK41 pKa = 9.04GQKK44 pKa = 9.33EE45 pKa = 4.23RR46 pKa = 11.84GSGTGYY52 pKa = 10.07EE53 pKa = 4.47HH54 pKa = 6.47YY55 pKa = 10.8QLVAAFAKK63 pKa = 10.08KK64 pKa = 10.04VSLPGVVGMFGRR76 pKa = 11.84GTHH79 pKa = 6.3GEE81 pKa = 4.04LSRR84 pKa = 11.84SEE86 pKa = 4.07AANEE90 pKa = 3.88YY91 pKa = 9.79VGKK94 pKa = 10.34EE95 pKa = 3.75EE96 pKa = 4.11TRR98 pKa = 11.84IGTTFEE104 pKa = 4.97LGAKK108 pKa = 9.57PIRR111 pKa = 11.84RR112 pKa = 11.84NSKK115 pKa = 8.97TDD117 pKa = 3.12WEE119 pKa = 4.97SVWTAAKK126 pKa = 10.54SGDD129 pKa = 3.9LSAVPANVRR138 pKa = 11.84VVNYY142 pKa = 9.29HH143 pKa = 6.17ALRR146 pKa = 11.84AIRR149 pKa = 11.84SDD151 pKa = 3.1HH152 pKa = 5.96AVPVGVEE159 pKa = 3.97RR160 pKa = 11.84TVHH163 pKa = 5.15VFWGSTGTGKK173 pKa = 10.02SRR175 pKa = 11.84RR176 pKa = 11.84AWEE179 pKa = 3.79EE180 pKa = 3.55AGIDD184 pKa = 4.16AYY186 pKa = 8.93TKK188 pKa = 10.48CPRR191 pKa = 11.84SKK193 pKa = 10.48FWDD196 pKa = 4.98GYY198 pKa = 10.19QDD200 pKa = 3.52QQHH203 pKa = 5.16VVVDD207 pKa = 4.2EE208 pKa = 4.15FRR210 pKa = 11.84GGIDD214 pKa = 3.25VAHH217 pKa = 7.59LLRR220 pKa = 11.84WFDD223 pKa = 3.83RR224 pKa = 11.84YY225 pKa = 9.38PVRR228 pKa = 11.84VEE230 pKa = 3.72IKK232 pKa = 9.65GSSRR236 pKa = 11.84PLVANTIWITSNKK249 pKa = 10.05DD250 pKa = 3.12PVEE253 pKa = 4.22WYY255 pKa = 9.79PEE257 pKa = 3.88LDD259 pKa = 3.67LATLEE264 pKa = 4.13ALLRR268 pKa = 11.84RR269 pKa = 11.84LTIVHH274 pKa = 6.7FPP276 pKa = 3.71
Molecular weight: 30.84 kDa
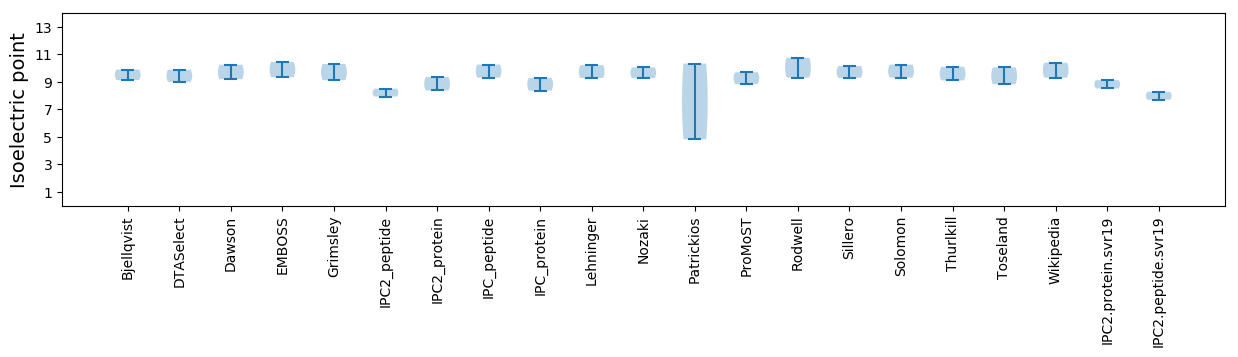
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GA81|A0A126GA81_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-41 OX=1685770 PE=4 SV=1
MM1 pKa = 7.27KK2 pKa = 10.42RR3 pKa = 11.84KK4 pKa = 9.22GHH6 pKa = 6.11YY7 pKa = 7.25PTPDD11 pKa = 2.88SSRR14 pKa = 11.84SRR16 pKa = 11.84DD17 pKa = 3.17RR18 pKa = 11.84KK19 pKa = 8.81RR20 pKa = 11.84AKK22 pKa = 9.93HH23 pKa = 5.61SRR25 pKa = 11.84PTIPSRR31 pKa = 11.84PLTHH35 pKa = 6.83RR36 pKa = 11.84VVQAHH41 pKa = 5.52TNYY44 pKa = 9.93INAIASRR51 pKa = 11.84YY52 pKa = 7.94NRR54 pKa = 11.84AVVGDD59 pKa = 3.8FLGGVPGAIIGAATSNVPFTIKK81 pKa = 9.35KK82 pKa = 8.5TIMGRR87 pKa = 11.84GNPAKK92 pKa = 10.63KK93 pKa = 9.94LVQRR97 pKa = 11.84SAMTSKK103 pKa = 10.77GKK105 pKa = 10.31NKK107 pKa = 10.22LKK109 pKa = 10.0TYY111 pKa = 10.45KK112 pKa = 10.02PYY114 pKa = 10.73KK115 pKa = 10.39LSDD118 pKa = 3.45KK119 pKa = 10.25FVKK122 pKa = 10.21SVKK125 pKa = 9.85QVLQTTCSTGTYY137 pKa = 7.21VTVRR141 pKa = 11.84QGLVGTLMNTAASGSMIGNDD161 pKa = 3.98LGQTGINQIWYY172 pKa = 8.28PGSSNPAGSRR182 pKa = 11.84TLFNALATLSDD193 pKa = 3.65AAATDD198 pKa = 3.88STVVAQTGLNFFTPAKK214 pKa = 9.81IIDD217 pKa = 3.87AASVLFNNKK226 pKa = 9.22GLGDD230 pKa = 4.25PYY232 pKa = 11.36SSAGNLATEE241 pKa = 5.01FTTADD246 pKa = 3.28GSIPTSNTAAGALKK260 pKa = 10.48INVLKK265 pKa = 10.84SYY267 pKa = 11.59VDD269 pKa = 4.48FEE271 pKa = 4.59MKK273 pKa = 10.49NLSNRR278 pKa = 11.84VAYY281 pKa = 10.36VDD283 pKa = 3.03IWEE286 pKa = 4.86CSPTLKK292 pKa = 10.41FQNINPLQVLQQTAAVWDD310 pKa = 3.87STSTINRR317 pKa = 11.84NITYY321 pKa = 7.99ATANANPNEE330 pKa = 4.12IFLDD334 pKa = 3.82STVDD338 pKa = 3.36GMGVLKK344 pKa = 10.42KK345 pKa = 10.72YY346 pKa = 10.69AGLPMSWKK354 pKa = 9.32KK355 pKa = 10.86RR356 pKa = 11.84SMVLAPDD363 pKa = 3.97EE364 pKa = 4.74TCLHH368 pKa = 6.15TVQGPKK374 pKa = 10.66GILDD378 pKa = 3.39WSKK381 pKa = 10.96LVSTQPSVAPATGQVNINLNCLLKK405 pKa = 10.51GFSVGCVISVRR416 pKa = 11.84TDD418 pKa = 2.99QVFQSGIPMYY428 pKa = 10.3GAKK431 pKa = 8.52KK432 pKa = 8.85TYY434 pKa = 10.71NDD436 pKa = 3.91GANLRR441 pKa = 11.84MAVPVAIEE449 pKa = 3.93MKK451 pKa = 10.0EE452 pKa = 4.15SYY454 pKa = 9.95KK455 pKa = 10.77VAVPEE460 pKa = 3.92VAGFVSRR467 pKa = 11.84AVVGGSAQPLNLRR480 pKa = 11.84KK481 pKa = 9.41PRR483 pKa = 11.84FVLWNQVNANGTTNVNKK500 pKa = 10.51LYY502 pKa = 10.74LSNEE506 pKa = 4.1LNPVYY511 pKa = 9.9DD512 pKa = 3.69TTQAQGG518 pKa = 3.0
MM1 pKa = 7.27KK2 pKa = 10.42RR3 pKa = 11.84KK4 pKa = 9.22GHH6 pKa = 6.11YY7 pKa = 7.25PTPDD11 pKa = 2.88SSRR14 pKa = 11.84SRR16 pKa = 11.84DD17 pKa = 3.17RR18 pKa = 11.84KK19 pKa = 8.81RR20 pKa = 11.84AKK22 pKa = 9.93HH23 pKa = 5.61SRR25 pKa = 11.84PTIPSRR31 pKa = 11.84PLTHH35 pKa = 6.83RR36 pKa = 11.84VVQAHH41 pKa = 5.52TNYY44 pKa = 9.93INAIASRR51 pKa = 11.84YY52 pKa = 7.94NRR54 pKa = 11.84AVVGDD59 pKa = 3.8FLGGVPGAIIGAATSNVPFTIKK81 pKa = 9.35KK82 pKa = 8.5TIMGRR87 pKa = 11.84GNPAKK92 pKa = 10.63KK93 pKa = 9.94LVQRR97 pKa = 11.84SAMTSKK103 pKa = 10.77GKK105 pKa = 10.31NKK107 pKa = 10.22LKK109 pKa = 10.0TYY111 pKa = 10.45KK112 pKa = 10.02PYY114 pKa = 10.73KK115 pKa = 10.39LSDD118 pKa = 3.45KK119 pKa = 10.25FVKK122 pKa = 10.21SVKK125 pKa = 9.85QVLQTTCSTGTYY137 pKa = 7.21VTVRR141 pKa = 11.84QGLVGTLMNTAASGSMIGNDD161 pKa = 3.98LGQTGINQIWYY172 pKa = 8.28PGSSNPAGSRR182 pKa = 11.84TLFNALATLSDD193 pKa = 3.65AAATDD198 pKa = 3.88STVVAQTGLNFFTPAKK214 pKa = 9.81IIDD217 pKa = 3.87AASVLFNNKK226 pKa = 9.22GLGDD230 pKa = 4.25PYY232 pKa = 11.36SSAGNLATEE241 pKa = 5.01FTTADD246 pKa = 3.28GSIPTSNTAAGALKK260 pKa = 10.48INVLKK265 pKa = 10.84SYY267 pKa = 11.59VDD269 pKa = 4.48FEE271 pKa = 4.59MKK273 pKa = 10.49NLSNRR278 pKa = 11.84VAYY281 pKa = 10.36VDD283 pKa = 3.03IWEE286 pKa = 4.86CSPTLKK292 pKa = 10.41FQNINPLQVLQQTAAVWDD310 pKa = 3.87STSTINRR317 pKa = 11.84NITYY321 pKa = 7.99ATANANPNEE330 pKa = 4.12IFLDD334 pKa = 3.82STVDD338 pKa = 3.36GMGVLKK344 pKa = 10.42KK345 pKa = 10.72YY346 pKa = 10.69AGLPMSWKK354 pKa = 9.32KK355 pKa = 10.86RR356 pKa = 11.84SMVLAPDD363 pKa = 3.97EE364 pKa = 4.74TCLHH368 pKa = 6.15TVQGPKK374 pKa = 10.66GILDD378 pKa = 3.39WSKK381 pKa = 10.96LVSTQPSVAPATGQVNINLNCLLKK405 pKa = 10.51GFSVGCVISVRR416 pKa = 11.84TDD418 pKa = 2.99QVFQSGIPMYY428 pKa = 10.3GAKK431 pKa = 8.52KK432 pKa = 8.85TYY434 pKa = 10.71NDD436 pKa = 3.91GANLRR441 pKa = 11.84MAVPVAIEE449 pKa = 3.93MKK451 pKa = 10.0EE452 pKa = 4.15SYY454 pKa = 9.95KK455 pKa = 10.77VAVPEE460 pKa = 3.92VAGFVSRR467 pKa = 11.84AVVGGSAQPLNLRR480 pKa = 11.84KK481 pKa = 9.41PRR483 pKa = 11.84FVLWNQVNANGTTNVNKK500 pKa = 10.51LYY502 pKa = 10.74LSNEE506 pKa = 4.1LNPVYY511 pKa = 9.9DD512 pKa = 3.69TTQAQGG518 pKa = 3.0
Molecular weight: 55.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
794 |
276 |
518 |
397.0 |
43.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.816 ± 0.511 | 1.134 ± 0.191 |
3.904 ± 0.049 | 3.526 ± 2.03 |
3.023 ± 0.144 | 8.312 ± 0.67 |
1.637 ± 0.763 | 4.408 ± 0.256 |
6.297 ± 0.741 | 7.431 ± 0.111 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.763 ± 0.628 | 5.919 ± 1.827 |
5.164 ± 0.274 | 3.526 ± 0.599 |
5.542 ± 1.469 | 7.431 ± 0.769 |
7.935 ± 0.854 | 9.068 ± 0.432 |
2.015 ± 0.972 | 3.149 ± 0.151 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
