
Chimpanzee faeces associated microphage 1
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
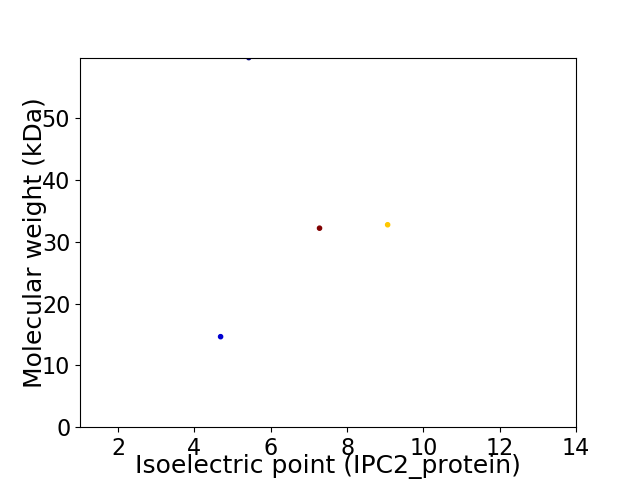
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A186YBN9|A0A186YBN9_9VIRU Minor CP 2 OS=Chimpanzee faeces associated microphage 1 OX=1676181 PE=4 SV=1
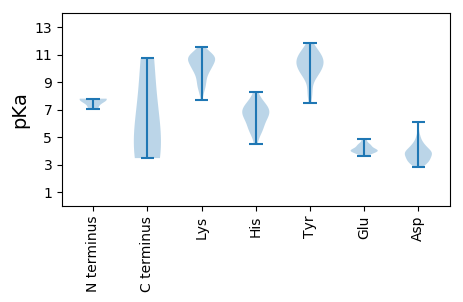
MM1 pKa = 7.79KK2 pKa = 10.5FFTLFDD8 pKa = 3.73KK9 pKa = 10.74PSPVGTACNTPSLTVQSEE27 pKa = 4.12KK28 pKa = 10.92AQCDD32 pKa = 3.02INAIIARR39 pKa = 11.84YY40 pKa = 9.21KK41 pKa = 8.51KK42 pKa = 9.09TGVVDD47 pKa = 4.59HH48 pKa = 6.74IQRR51 pKa = 11.84GEE53 pKa = 3.88PLYY56 pKa = 10.91ADD58 pKa = 4.42CEE60 pKa = 4.11QAITDD65 pKa = 4.07LAKK68 pKa = 10.83ARR70 pKa = 11.84ILVEE74 pKa = 4.09DD75 pKa = 4.31TEE77 pKa = 4.65DD78 pKa = 4.25AFWTLPSSVRR88 pKa = 11.84DD89 pKa = 3.92VIGEE93 pKa = 3.98PSNLPSWASANRR105 pKa = 11.84SEE107 pKa = 4.05AEE109 pKa = 4.03KK110 pKa = 11.15YY111 pKa = 10.94GFLAPQATPPADD123 pKa = 3.42SVTPPPTDD131 pKa = 3.11SAANGG136 pKa = 3.47
MM1 pKa = 7.79KK2 pKa = 10.5FFTLFDD8 pKa = 3.73KK9 pKa = 10.74PSPVGTACNTPSLTVQSEE27 pKa = 4.12KK28 pKa = 10.92AQCDD32 pKa = 3.02INAIIARR39 pKa = 11.84YY40 pKa = 9.21KK41 pKa = 8.51KK42 pKa = 9.09TGVVDD47 pKa = 4.59HH48 pKa = 6.74IQRR51 pKa = 11.84GEE53 pKa = 3.88PLYY56 pKa = 10.91ADD58 pKa = 4.42CEE60 pKa = 4.11QAITDD65 pKa = 4.07LAKK68 pKa = 10.83ARR70 pKa = 11.84ILVEE74 pKa = 4.09DD75 pKa = 4.31TEE77 pKa = 4.65DD78 pKa = 4.25AFWTLPSSVRR88 pKa = 11.84DD89 pKa = 3.92VIGEE93 pKa = 3.98PSNLPSWASANRR105 pKa = 11.84SEE107 pKa = 4.05AEE109 pKa = 4.03KK110 pKa = 11.15YY111 pKa = 10.94GFLAPQATPPADD123 pKa = 3.42SVTPPPTDD131 pKa = 3.11SAANGG136 pKa = 3.47
Molecular weight: 14.64 kDa
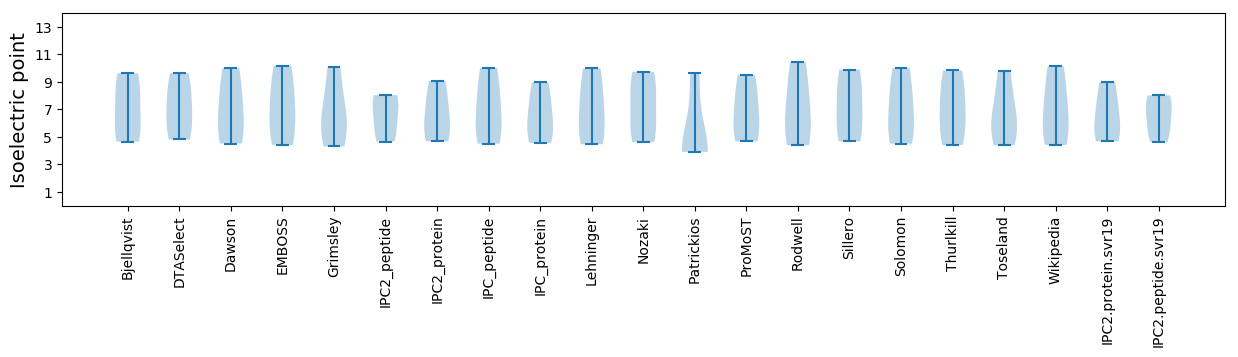
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A186YBP1|A0A186YBP1_9VIRU Major CP OS=Chimpanzee faeces associated microphage 1 OX=1676181 PE=3 SV=1
MM1 pKa = 7.78AFGIDD6 pKa = 4.05DD7 pKa = 4.28ALMAAVAGGTQGAMNLGGAVLGDD30 pKa = 3.49YY31 pKa = 10.52YY32 pKa = 11.37NRR34 pKa = 11.84KK35 pKa = 8.98AASKK39 pKa = 8.81QNKK42 pKa = 8.73VSWEE46 pKa = 3.8MMLANQQWLEE56 pKa = 4.03RR57 pKa = 11.84MSNTAHH63 pKa = 5.48QRR65 pKa = 11.84EE66 pKa = 4.52VADD69 pKa = 4.56LRR71 pKa = 11.84AAGLNPILSATGGSGASSPSYY92 pKa = 10.28AASAPDD98 pKa = 4.02LSALGKK104 pKa = 10.27SGEE107 pKa = 4.23GFSRR111 pKa = 11.84VGNGVMDD118 pKa = 4.01KK119 pKa = 10.9ALRR122 pKa = 11.84AASLRR127 pKa = 11.84SEE129 pKa = 4.43LKK131 pKa = 10.94LMEE134 pKa = 4.85SNANNAEE141 pKa = 3.94AQAANSWEE149 pKa = 3.87NVLYY153 pKa = 10.44TKK155 pKa = 11.16ANTAKK160 pKa = 9.96VLAEE164 pKa = 3.95AGVFQSQLDD173 pKa = 3.88RR174 pKa = 11.84IGWLKK179 pKa = 10.81KK180 pKa = 10.12NSPNTWYY187 pKa = 10.63LWGDD191 pKa = 3.34KK192 pKa = 10.7DD193 pKa = 3.72PSVWQLLPGFSRR205 pKa = 11.84GSEE208 pKa = 3.94FGAQSVGNGVKK219 pKa = 9.88SVYY222 pKa = 10.77DD223 pKa = 3.61FVKK226 pKa = 10.73DD227 pKa = 4.53PIASAVKK234 pKa = 9.95KK235 pKa = 10.48VYY237 pKa = 10.6NAVKK241 pKa = 10.73DD242 pKa = 4.81FGANNASSSSNQSIGYY258 pKa = 9.26SSYY261 pKa = 11.84DD262 pKa = 3.23KK263 pKa = 11.21GSSSDD268 pKa = 3.42SGKK271 pKa = 10.32VPLTTTRR278 pKa = 11.84WRR280 pKa = 11.84KK281 pKa = 8.19GQPLRR286 pKa = 11.84SGKK289 pKa = 9.74FDD291 pKa = 3.5YY292 pKa = 11.27QEE294 pKa = 3.79VRR296 pKa = 11.84RR297 pKa = 11.84KK298 pKa = 10.06RR299 pKa = 11.84LEE301 pKa = 3.92NLKK304 pKa = 10.57KK305 pKa = 10.71
MM1 pKa = 7.78AFGIDD6 pKa = 4.05DD7 pKa = 4.28ALMAAVAGGTQGAMNLGGAVLGDD30 pKa = 3.49YY31 pKa = 10.52YY32 pKa = 11.37NRR34 pKa = 11.84KK35 pKa = 8.98AASKK39 pKa = 8.81QNKK42 pKa = 8.73VSWEE46 pKa = 3.8MMLANQQWLEE56 pKa = 4.03RR57 pKa = 11.84MSNTAHH63 pKa = 5.48QRR65 pKa = 11.84EE66 pKa = 4.52VADD69 pKa = 4.56LRR71 pKa = 11.84AAGLNPILSATGGSGASSPSYY92 pKa = 10.28AASAPDD98 pKa = 4.02LSALGKK104 pKa = 10.27SGEE107 pKa = 4.23GFSRR111 pKa = 11.84VGNGVMDD118 pKa = 4.01KK119 pKa = 10.9ALRR122 pKa = 11.84AASLRR127 pKa = 11.84SEE129 pKa = 4.43LKK131 pKa = 10.94LMEE134 pKa = 4.85SNANNAEE141 pKa = 3.94AQAANSWEE149 pKa = 3.87NVLYY153 pKa = 10.44TKK155 pKa = 11.16ANTAKK160 pKa = 9.96VLAEE164 pKa = 3.95AGVFQSQLDD173 pKa = 3.88RR174 pKa = 11.84IGWLKK179 pKa = 10.81KK180 pKa = 10.12NSPNTWYY187 pKa = 10.63LWGDD191 pKa = 3.34KK192 pKa = 10.7DD193 pKa = 3.72PSVWQLLPGFSRR205 pKa = 11.84GSEE208 pKa = 3.94FGAQSVGNGVKK219 pKa = 9.88SVYY222 pKa = 10.77DD223 pKa = 3.61FVKK226 pKa = 10.73DD227 pKa = 4.53PIASAVKK234 pKa = 9.95KK235 pKa = 10.48VYY237 pKa = 10.6NAVKK241 pKa = 10.73DD242 pKa = 4.81FGANNASSSSNQSIGYY258 pKa = 9.26SSYY261 pKa = 11.84DD262 pKa = 3.23KK263 pKa = 11.21GSSSDD268 pKa = 3.42SGKK271 pKa = 10.32VPLTTTRR278 pKa = 11.84WRR280 pKa = 11.84KK281 pKa = 8.19GQPLRR286 pKa = 11.84SGKK289 pKa = 9.74FDD291 pKa = 3.5YY292 pKa = 11.27QEE294 pKa = 3.79VRR296 pKa = 11.84RR297 pKa = 11.84KK298 pKa = 10.06RR299 pKa = 11.84LEE301 pKa = 3.92NLKK304 pKa = 10.57KK305 pKa = 10.71
Molecular weight: 32.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1253 |
136 |
537 |
313.3 |
34.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.46 ± 1.653 | 1.676 ± 0.749 |
6.385 ± 0.493 | 4.389 ± 0.805 |
4.549 ± 0.679 | 7.502 ± 0.836 |
1.756 ± 0.676 | 3.033 ± 0.65 |
4.709 ± 0.987 | 8.22 ± 0.602 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.474 ± 0.472 | 4.868 ± 0.834 |
5.108 ± 0.951 | 3.352 ± 0.537 |
6.065 ± 0.947 | 9.417 ± 1.088 |
5.666 ± 1.19 | 6.145 ± 0.349 |
2.314 ± 0.171 | 3.911 ± 0.443 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
